Abstract
Background
Osteoporosis is the most common metabolic bone disease and can cause fragility fractures. Despite this, screening utilization rates for osteoporosis remain low among populations at risk. Automated bone mineral density (BMD) estimation using computed tomography (CT) can help bridge this gap and serve as an alternative screening method to dual-energy X-ray absorptiometry (DXA).
Methods
The feasibility of an opportunistic and population agnostic screening method for osteoporosis using abdominal CT scans without bone densitometry phantom-based calibration was investigated in this retrospective study. A total of 268 abdominal CT-DXA pairs and 99 abdominal CT studies without DXA scores were obtained from an oncology specialty clinic in the Republic of Korea. The center axial CT slices from the L1, L2, L3, and L4 lumbar vertebrae were annotated with the CT slice level and spine segmentation labels for each subject. Deep learning models were trained to localize the center axial slice from the CT scan of the torso, segment the vertebral bone, and estimate BMD for the top four lumbar vertebrae.
Osteoporosis is a common condition that causes bone fragility. With increasing life expectancy and a graying population across the world, more individuals are projected to be at risk for this condition [1,2]. Osteoporotic fractures are associated with a significantly increased risk of long-term disability [3] and mortality [4,5]. In the United States, approximately 21% of women and 32% of men aged 65 or older die within 1 year of a hip fracture [6]. Although the condition is both preventable and treatable with early detection as well as its associated socioeconomic burden, osteoporosis remains underdiagnosed [7].
The gold standard modality for estimating bone mineral density (BMD) and diagnosing osteoporosis is dual-energy X-ray absorptiometry (DXA) [8-10]. However, timely diagnosis of this condition is difficult as osteoporosis is asymptomatic [10], and individuals at risk of fracture often have other comorbidities that require more immediate care [11]. To address this disconnection between disease prevalence and diagnosis, previous studies proposed manual and machine learning-based opportunistic osteoporosis screening methods using computed tomography (CT) as an alternative to DXA [12-14]. These methods do not require additional radiation, cost, or, if completely automated, input from health professionals. However, as these systems do not generate end-to-end predictions [15,16] or population agnostic BMD using DXA (BMDDXA) estimates [17-20], there are limitations in applying these results to real-world clinical practice.
This study proposes an automated, end-to-end, deep learning-based method for opportunistic osteoporosis screening of the top four lumbar vertebrae using abdominal CT scans. In contrast to the results of previous studies on CT-based osteoporosis screening, our system generates BMDDXA estimates that are population agnostic and do not rely on calibration using bone densitometry phantoms.
This retrospective study was approved by the Institutional Review Board of the National Cancer Center in the Republic of Korea (NCC2020-0165), and all identifying information was anonymized before analysis. The data used in this study included 367 (198 females and 169 males) contrast-enhanced abdominal CT scans of patients who underwent routine cancer screening between 2010 and 2019 at National Cancer Center in the Republic of Korea. Of these subjects, 268 had associated lumbar DXA values collected within 180 days of their CT scan, irrespective of the imaging order. Patients with spinal implants or DXA z-scores greater than 3.3 or less than –3.3 were excluded from this study. We did not include cases with a suspected fracture on CT scout films or a suspected fracture on DXA with a T-score difference of more than 1 between adjacent vertebrae.
Table 1 contains additional demographic information on the DXA subject population. Subjects were grouped into normal, osteopenia, and osteoporosis groups using their L1–L4 DXA T-score values and the World Health Organization (WHO) diagnostic criteria. Body mass index (BMI) status was based on the WHO BMI cutoffs for Asian populations, where a value below or equal to 18.5 is underweight, between 18.5 and 23 is normal, between 23 and 27.5 is overweight, and greater than 27.5 is obese [21].
The CTs and Convolution Kernels used were: (1) Discovery CT 750HD with Convolution Kernel: Standard (GE Healthcare, Waukesha, WI, USA) (n=238); (2) Brilliance with Convolution Kernel: B (Philips Medical Systems, Best, the Netherlands) (n=109); (3) SOMATOM Definition Edge with Convolution Kernel: B30f (Siemens Healthcare, Forchheim, Germany) (n=20). All CTs were scanned 80 seconds after injection of Omnipaqu (Iohexol, GE Healthcare) 300 contrast medium (Venous phase). Peak tube voltage was 120 kVp (n=347) or 100 kVp (n=20) The tube voltage used for all abdominal CT images was 120 kvP and the tube current (milliampere) was adjusted automatically by the thickness of the patient’s body for radiation dose optimization.
Lumbar DXA scans were performed using QDR 4,500 W (n=165), Horizon W (n=98), and Discovery W (n=5) scanners manufactured by Hologic (Marlborough, MA, USA). For each subject, slice number annotations and vertebral segmentation mask labels were assigned to the central axial CT slice of the L1, L2, L3, and L4 vertebrae.
Fig. 1 illustrates the CT-based opportunistic osteoporosis screening method. The system generated end-to-end BMD using convolutional neural network (BMDCNN) estimates from abdominal CT scans via three successive steps: localization, segmentation, and estimation. The localization subtask takes the abdominal CT scan of a subject as the input and outputs the center axial CT slice locations for the L1, L2, L3, and L4 vertebrae as integer values. For each of the four predicted center slices, the system takes one slice above and two slices below the detected center, segmenting the vertebral bone into four slices during the segmentation step. The segmented slices corresponding to each lumbar vertebra were cropped, concatenated counterclockwise, and used as the input for the estimation step to generate the BMDCNN predictions.
The localization subtask uses deep learning-based regressors to detect the center axial slice locations of the L1, L2, L3, and L4 spines. Our localization method is a two-step process that utilizes both frontal and sagittal maximum intensity projections (MIPs) and two regression models. In contrast to the previous work on CT-based lumbar spine localization by Belharbi et al. [22] and Kanavati et al. [23], our method outputs vertebral-level predictions for the top four lumbar vertebrae rather than just the center L3 slice location. Fig. 2 shows the overall flow of the proposed method.
The two stages of the localization method are trained separately and combined during the inference phase. In the first stage, a neural network outputs the top axial slice location of the L1 vertebra using frontal MIP, and the second model predicts the center slice locations of the L1, L2, L3, and L4 vertebrae during the second stage. The frontal MIP used in the first stage was center cropped along the x-axis and zero padded along the y-axis to generate a 160×256 pixel image. In the second stage, the sagittal MIP was cropped from the 120th pixel to the 280th pixel along the x-axis and 80 slices from the top L1 slice along the y-axis to generate a 160×80-pixel region of interest. During training, the sagittal MIP was cropped, starting from a random slice between 5 and 15 above the top L1 location ground-truth, and the MIP was cropped from 10 slices above the top L1 slice prediction in the first step of the localization subtask. Both stages use VGG19-based regressors [24], with a channel attention block applied before each pooling layer. Additional implementation details are outlined in Supplemental Fig. S1.
Spine BMDDXA measurements are known to be affected by body composition [25]; therefore, our system segments the vertebra for each axial CT slice using a U-Net-based model [26], such that the subsequent estimation subtask will only use bone images as input. However, there is a trade-off between improving model performance with more labeled samples and the resource demands associated with manual data annotation. To address this disconnection, we take a semi-supervised approach for this subtask, where we only use one segmentation mask for each vertebra but use all available axial slices during training via the Mumford-Shah (MS) loss [27]. As shown in Fig. 3, a semi-supervised learning approach allows us to use all the CT slices in a vertebra, leading to an approximately ten-fold increase in training data as compared to a strongly supervised approach using only the center vertebral slices. The loss was calculated using a weighted sum of cross-entropy (CE) and MS losses, where the CE loss was only applied to samples with ground-truth segmentation masks, whereas the MS loss was applied to all training samples. Additional details on the model architecture, loss function, and training procedure are provided in Supplemental Fig. S2.
In this study, BMD estimation was considered as an image regression task, where the system generated continuous BMDCNN predictions for the L1, L2, L3, and L4 vertebrae independently using a DenseNet169-based regressor [28]. Fig. 4 illustrates the image generation process for the BMD estimation subtask. For training, all axial CT slices from the entire range of a given vertebra were segmented, whereas the images used for testing were based on the output predictions from the detection and segmentation subtasks. For a given vertebra, four consecutive axial CT slices were selected to generate one input sample. During training and validation, a sliding window was applied to the slices, such that for a vertebra with n ordered slices, n−3 samples containing four adjacent CT slices were generated. In contrast to using only the slices near the center, this approach uses data from the entire vertebra during training while ensuring that slices closer to the start and end of the vertebra, which contain a higher proportion of denser cortical bone, are sampled less frequently. During testing, the center slice prediction for a given vertebra was used from the detection step, and one slice above and two slices below the predicted slice were selected. This choice, in contrast to two slices above and one slice below the detected center, was arbitrary because the differences between the two methods were negligible during model selection. Additional training and architecture details for the estimation subtask are available in Supplemental Methods.
The models in this study were evaluated using Dice scores, regression metrics (mean absolute error [MAE], root mean squared error, and mean absolute percentage error), Pearson correlation coefficients, and coefficients of determination. Dice scores were calculated using PyTorch (https://pytorch.org) [29]. Regression metrics and coefficients of determination were evaluated using Python and the scikit-learn [30] package. Pearson r coefficients and their respective P values were acquired using Python and the SciPy [31] package.
Table 2 presents the results of the proposed localization method at the vertebral and subject levels. Localization performance was evaluated in CT slices with an absolute error. The performance with the channel attention block was listed in Supplemental Table S1.
For each CT slice, segmentation maps were generated using the largest connected component of the predicted output. Predictions were evaluated using Dice scores. For a segmentation map A and ground-truth label B, the Dice score is defined as:
where |A∩B| is the area of the overlapping region between A and B and |A|+|B| is the sum of the areas A and B. Since the proposed screening method is performed end-to-end, segmentation performance is contingent on the results of the localization subtask (Supplemental Fig. S3). However, the vertebral segmentation masks were evaluated and the Dice scores on the manually labeled center vertebral slices were computed. Table 3 reports the results of our semi-supervised segmentation method at the subject and vertebral levels compared with a standard, strongly supervised U-Net. The detailed results were presented in Supplemental Table S2.
Table 4 lists the end-to-end performance of the estimation subtask. The 98 subjects in the test set (52 females and 46 males) were represented by 392 image samples generated from 1,568 CT slices, where each image represented one lumbar vertebra. Vertebral-level results were obtained by evaluating the end-toend BMDCNN estimates against the BMDDXA ground truths, while the subject-level results were generated by taking the arithmetic mean of the L1, L2, L3, and L4 BMDCNN predictions and comparing them to the total lumbar BMDDXA values. Fig. 5 illustrates these results using regression and Bland-Altman [32] plots.
This study proposes a deep learning-based system for end-to-end opportunistic osteoporosis screening using abdominal CT scans. Our system detects the center L1, L2, L3, and L4 axial CT slices with a MAE of 1.07±2.31 slices, segments the vertebra with a Dice score of 0.968, and generates vertebra-level BMDDXA estimates with a MAE of 0.079, Pearson r of 0.852 (P<0.001), and R2 of 0.714 on the held-out test set. For subject-level predictions, the system output predictions had an MAE of 0.066, Pearson r of 0.907 (P<0.001), and R2 of 0.781.
Previous studies have investigated deep learning-based methods for CT-based BMD estimation. Some of these studies relied on extracting features on manually selected CT slices rather than entire studies. Tang et al. [16] proposed a method of screening for osteoporosis using a multiclass classification network to discriminate between normal bone mass, low bone mass, and osteoporosis from segmented axial CT slices, while Yasaka et al. [15] used manually cropped mid-vertebral axial CT slices with circular region-of-interest markers to train a convolutional neural network (CNN) to output continuous BMDDXA predictions. Other more recent studies took end-to-end approaches to BMD estimation. Pickhardt et al. [33] and Fang et al. [18] presented end-to-end automatic methods to predict quantitative CT (QCT) values, while Liu et al. [19] correlated the mean Hounsfield unit values in regions of interest from low-dose chest CT scans with BMDDXA values for the thoracic and first two lumbar vertebrae. Krishnaraj et al. [20] used a cascade of two segmentation networks and machine learning-based regression to predict the DXA t-scores for the top four lumbar vertebrae from extracted voxel volumes. With the exception of Liu et al. [34] and Pickhardt et al. [33], these prior studies relied on strongly supervised methods (i.e., each training sample has a corresponding ground-truth label) for slice detection and region-of-interest extraction tasks. To reduce the cost associated with manual annotation, our work adopts a semi-supervised approach to segmentation, where only the ground-truth masks for the center axial CT slice of each vertebra are provided. We note that while the localization and segmentation approaches used by Liu et al. [34] and Pickhardt et al. [33] require no masks, the respective methods require anatomical landmarks (i.e., clavicles) that are not present in our dataset of abdominal CT scans [34] or the use of the lowest ribs [35], which can be difficult owing to lumbar or absent ribs.
Our system generates end-to-end predictions of BMDDXA values from abdominal CT scans rather than DXA t-scores [20], normal/osteopenia/osteoporosis classes [16], or QCT BMD values [17,18]. DXA t-scores and DXA-based osteoporosis classifications are population-dependent, and thresholds for osteopenia and osteoporosis do not always generalize across different groups. Although QCT has certain benefits over DXA, including capturing volumetric rather than areal BMD, the screening modality has lower utilization than DXA and often requires the use of a BMD calibration phantom at the time of the scan. This limits the number of available QCT samples relative to that of DXA, leaving fewer CT-BMD pairs for future training and validation.
This study has a few limitations of its own. All DXA and CT studies were sourced via routine cancer screening at a single oncology specialty center in the Republic of Korea, limiting the scope of our analysis as BMD varies between national and ethnic groups [2,36]. To generate vertebral-level annotations frontal MIP images were compared to lumbar DXA images. This was because of the difficulty introduced by the transitional vertebrae, lumbar ribs, and anatomical markers not being present in abdominal CTs, which would enable annotation with CT images alone. In particular, lumbosacral transitional vertebrae (LSTV)— common anatomical variants of the lumbar spine caused by the lumbarization of the S1 vertebra (leading to the appearance of six lumbar vertebrae) and sacralization of the L5 vertebra (leading to the appearance of four lumbar vertebrae)—make vertebral-level annotation difficult even for trained clinicians [37]. In practice, LSTVs cause errors in imaging [38] and wrong-site surgery [39]. To account for these variations, DXA images were used as ground-truth labels to reflect real-world clinical decision-making. Additionally, we were unable to collect information on family history, menopausal status, physical activity, or other risk factors.
This study describes an end-to-end deep learning-based system to measure BMD using abdominal CT. Furthermore, this study demonstrated that CT scans collected during routine cancer screening without bone densitometry phantoms could generate BMDDXA predictions without additional tests.
In this study, we have shown that machine learning can be used to predict BMD at each level of the lumbar spine from routine CT images. Next, we would like to develop a machine learning model that predicts the T-score and z-score of L1–L4 from CT images and develop software to diagnose osteoporosis. We also want to develop artificial intelligence software that uses clinical data from electronic health records and CT images to predict the long-term risk of fractures.
Supplementary Material
Supplemental Table S1.
Results of Spine Localization on 98 Test Data
Supplemental Table S2.
Results of Cross-Validation on 269 Training Data
Supplemental Fig. S1.
(A) The network architecture for the localization subtask models. (B) Channel attention block architecture. The number below each block denotes the number of channels. BN, batch norm; ReLU, rectified linear unit; Conv, convolution.
Supplemental Fig. S2.
(A) Proposed semi-supervised learning model for the lumbar segmentation method. (B) The architecture of the segmentation network. LCE, cross-entropy loss; Y, predicted segmentation map; LMScnn, Mumford-Shah loss; Conv, convolution; BN, batch norm; ReLU, rectified linear unit.
Supplemental Fig. S3.
Results of lumbar vertebra (L1, L2, L3, and L4) segmentation from the proposed method on the computed tomography scans of (A) a man and (B) a woman. Dice scores are notated in each result.
ACKNOWLEDGMENTS
This work was funded by Korea Advanced Institute of Science and Technology (KAIST) R&D Program (KI Meta-Convergence Program) 2020 (N10200012) through KAIST and by a grant from the National Cancer Center (2010080 and 2310840) in the Republic of Korea. Boah Kim is now at National Institutes of Health, Bethesda, MD, USA.
REFERENCES
1. Wright NC, Looker AC, Saag KG, Curtis JR, Delzell ES, Randall S, et al. The recent prevalence of osteoporosis and low bone mass in the United States based on bone mineral density at the femoral neck or lumbar spine. J Bone Miner Res. 2014; 29:2520–6.

2. Zeng Q, Li N, Wang Q, Feng J, Sun D, Zhang Q, et al. The prevalence of osteoporosis in China, a nationwide, multicenter DXA survey. J Bone Miner Res. 2019; 34:1789–97.

3. Dyer SM, Crotty M, Fairhall N, Magaziner J, Beaupre LA, Cameron ID, et al. A critical review of the long-term disability outcomes following hip fracture. BMC Geriatr. 2016; 16:158.

4. Alarkawi D, Bliuc D, Tran T, Ahmed LA, Emaus N, Bjornerem A, et al. Impact of osteoporotic fracture type and subsequent fracture on mortality: the Tromsø Study. Osteoporos Int. 2020; 31:119–30.

5. Haentjens P, Magaziner J, Colon-Emeric CS, Vanderschueren D, Milisen K, Velkeniers B, et al. Meta-analysis: excess mortality after hip fracture among older women and men. Ann Intern Med. 2010; 152:380–90.

6. Brauer CA, Coca-Perraillon M, Cutler DM, Rosen AB. Incidence and mortality of hip fractures in the United States. JAMA. 2009; 302:1573–9.

7. Miller PD. Underdiagnosis and undertreatment of osteoporosis: the battle to be won. J Clin Endocrinol Metab. 2016; 101:852–9.
8. Kanis JA. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: synopsis of a WHO report. WHO Study Group. Osteoporos Int. 1994; 4:368–81.
9. US Preventive Services Task Force; Curry SJ, Krist AH, Owens DK, Barry MJ, Caughey AB, et al. Screening for osteoporosis to prevent fractures: US Preventive Services Task Force recommendation statement. JAMA. 2018; 319:2521–31.
10. Cosman F, de Beur SJ, LeBoff MS, Lewiecki EM, Tanner B, Randall S, et al. Clinician’s guide to prevention and treatment of osteoporosis. Osteoporos Int. 2014; 25:2359–81.

12. Buckens CF, Dijkhuis G, de Keizer B, Verhaar HJ, de Jong PA. Opportunistic screening for osteoporosis on routine computed tomography?: an external validation study. Eur Radiol. 2015; 25:2074–9.

13. Jang S, Graffy PM, Ziemlewicz TJ, Lee SJ, Summers RM, Pickhardt PJ. Opportunistic osteoporosis screening at routine abdominal and thoracic CT: normative L1 trabecular attenuation values in more than 20,000 adults. Radiology. 2019; 291:360–7.

14. Dagan N, Elnekave E, Barda N, Bregman-Amitai O, Bar A, Orlovsky M, et al. Automated opportunistic osteoporotic fracture risk assessment using computed tomography scans to aid in FRAX underutilization. Nat Med. 2020; 26:77–82.

15. Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol. 2020; 30:3549–57.

16. Tang C, Zhang W, Li H, Li L, Li Z, Cai A, et al. CNN-based qualitative detection of bone mineral density via diagnostic CT slices for osteoporosis screening. Osteoporos Int. 2021; 32:971–9.

17. Pickhardt PJ, Graffy PM, Zea R, Lee SJ, Liu J, Sandfort V, et al. Automated abdominal CT imaging biomarkers for opportunistic prediction of future major osteoporotic fractures in asymptomatic adults. Radiology. 2020; 297:64–72.

18. Fang Y, Li W, Chen X, Chen K, Kang H, Yu P, et al. Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. Eur Radiol. 2021; 31:1831–42.

19. Liu S, Gonzalez J, Zulueta J, de Torres JP, Yankelevitz DF, Henschke CI, et al. Fully automated bone mineral density assessment from low-dose chest CT. In : Petrick N, Mori K, editors. Proceedings Volume 10575, SPIE Medical Imaging 2018: Computer-Aided Diagnosis; 2018 Feb 10-15; Houston, TX. Bellingham: International Society for Optics and Photonics;2018. Available from: https://doi.org/10.1117/12.2293838.

20. Krishnaraj A, Barrett S, Bregman-Amitai O, Cohen-Sfady M, Bar A, Chettrit D, et al. Simulating dual-energy X-ray absorptiometry in CT using deep-learning segmentation cascade. J Am Coll Radiol. 2019; 16:1473–9.

21. WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004; 363:157–63.
22. Belharbi S, Chatelain C, Herault R, Adam S, Thureau S, Chastan M, et al. Spotting L3 slice in CT scans using deep convolutional network and transfer learning. Comput Biol Med. 2017; 87:95–103.

23. Kanavati F, Islam S, Aboagye EO, Rockall A. Automatic L3 slice detection in 3D CT images using fully-convolutional networks. arXiv [Preprint]. 2018; Nov. 22. . https://doi.org/10.48550/arXiv.1811.09244.

24. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. 2015; Apr. 10. https://doi.org/10.48550/arXiv.1409.1556.

25. Yu EW, Thomas BJ, Brown JK, Finkelstein JS. Simulated increases in body fat and errors in bone mineral density measurements by DXA and QCT. J Bone Miner Res. 2012; 27:119–24.

26. Navab N, Hornegger J, Wells WM, Frangi A. Medical Image Computing and Computer-Assisted Intervention: MICCAI 2015. Lecture notes in computer science. Vol. 9351. Cham: Springer;2015. Chapter, U-net: convolutional networks for biomedical image segmentation. 234–41. [cited 2024 Mar 16]. Available from: https://doi.org/10.1007/978-3-319-24574-4_28.

27. Kim B, Ye JC. Mumford-Shah loss functional for image segmentation with deep learning. IEEE Trans Image Process. 2020; 29:1856–66.

28. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In : Proceedings of the 2017 IEEE conference on computer vision and pattern recognition (CVPR); 2017 Jul 21-26; Honolulu, HI. Washington DC: IEEE Computer Society;2017. p. 2261–9. Available from: https://doi.org/10.1109/CVPR.2017.243.

29. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019; 32:1–12.
30. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12:2825–30.
31. Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020; 17:261–72.
32. Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. J R Stat Soc Series D Stat. 1983; 32:307–17.

33. Pickhardt PJ, Lee SJ, Liu J, Yao J, Lay N, Graffy PM, et al. Population-based opportunistic osteoporosis screening: validation of a fully automated CT tool for assessing longitudinal BMD changes. Br J Radiol. 2019; 92:20180726.

34. Liu S, Xie Y, Reeves AP. Individual bone structure segmentation and labeling from low-dose chest CT. In : Armato SG, Petrick NA, editors. Proceedings Volume 10134, Medical Imaging 2017: Computer-Aided Diagnosis; 2017 Feb 11-16; Orlando. FL. Bellingham: International Society for Optics and Photonics;2017. Available from: https://doi.org/10.1117/12.2254162.

35. Summers RM, Baecher N, Yao J, Liu J, Pickhardt PJ, Choi JR, et al. Feasibility of simultaneous computed tomographic colonography and fully automated bone mineral densitometry in a single examination. J Comput Assist Tomogr. 2011; 35:212–6.

36. Nam HS, Kweon SS, Choi JS, Zmuda JM, Leung PC, Lui LY, et al. Racial/ethnic differences in bone mineral density among older women. J Bone Miner Metab. 2013; 31:190–8.

37. Jancuska JM, Spivak JM, Bendo JA. A review of symptomatic lumbosacral transitional vertebrae: Bertolotti’s syndrome. Int J Spine Surg. 2015; 9:42.

Fig. 1.
Overview of the end-to-end opportunistic osteoporosis screening method used. The grey text boxes denote deep learning-based subtasks. (A) The abdominal computed tomography (CT) scan of a subject is input into the system. (B) Both frontal and sagittal maximum intensity projections are generated from the CT scan. (C) Two deep learning-based regressors predict the center axial slice locations for the top four lumbar vertebrae. (D) For each of the four vertebrae, one slice above and two slices below the respective center slice prediction are selected. (E) Vertebral bone, including the spinous process, is segmented with a trained model. (F) Using the segmentation masks from the previous step, a 196×196-pixel area containing the segmented vertebral bone is cropped from each slice. For each of the four lumbar vertebrae, the four selected slices are concatenated counterclockwise to form a 392×392-pixel image to be used as inputs for the estimation algorithm. (G) An image regressor generates bone mineral density (BMD) using convolutional neural network BMD estimates for the four vertebrae independently.
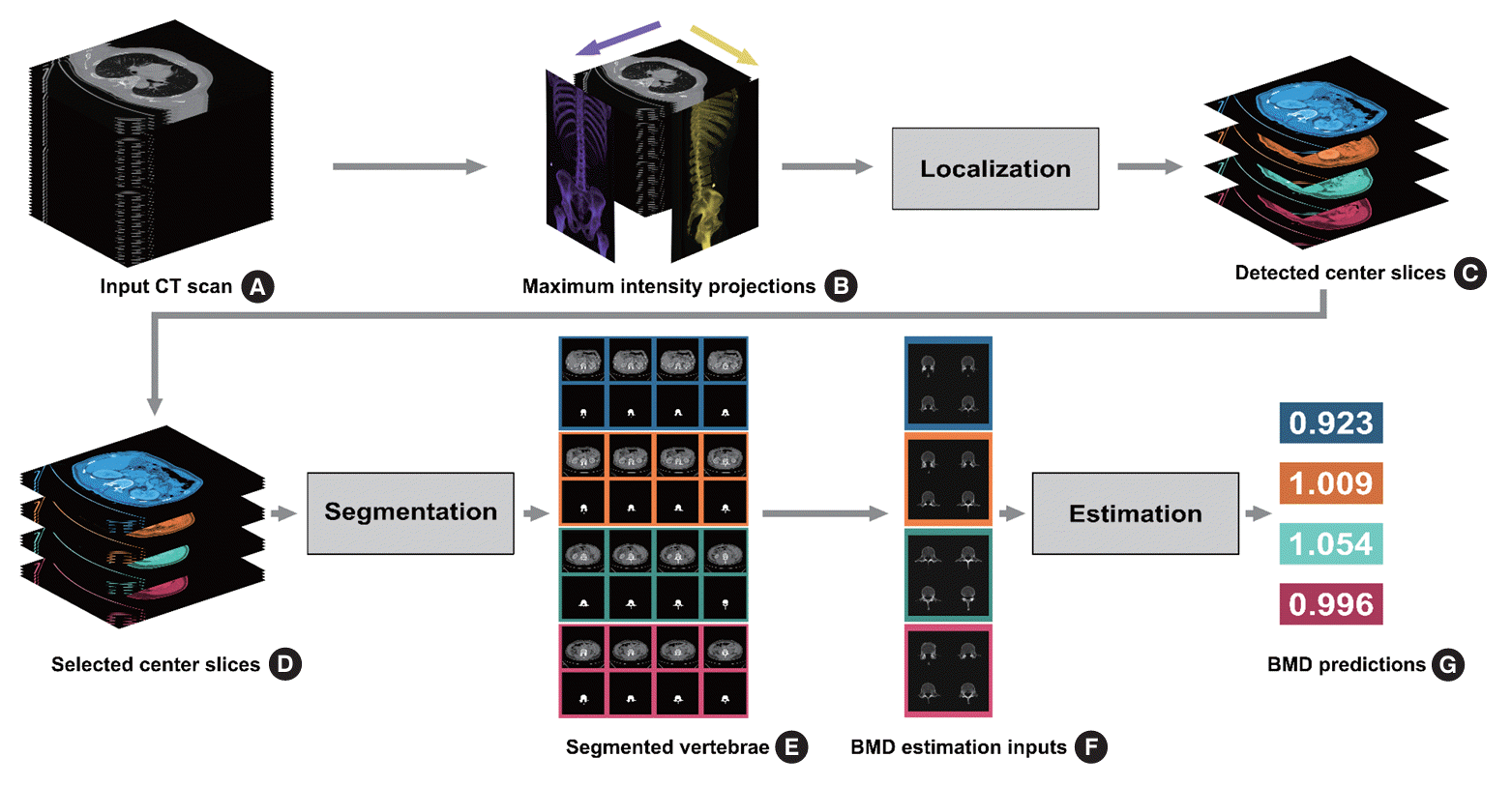
Fig. 2.
Overall flow of the proposed lumbar spine localization method. (A) In the first stage, deep neural network 1 predicts the index of the top axial slice of the l1 vertebra. (B) During the second stage, deep neural network 2 outputs predictions for the center slice for the l1, l2, l3, and l4 vertebrae. The numbers below each image denote image dimensions. 3D, three-dimensional; CT, computed tomography; MIP, maximum intensity projection.
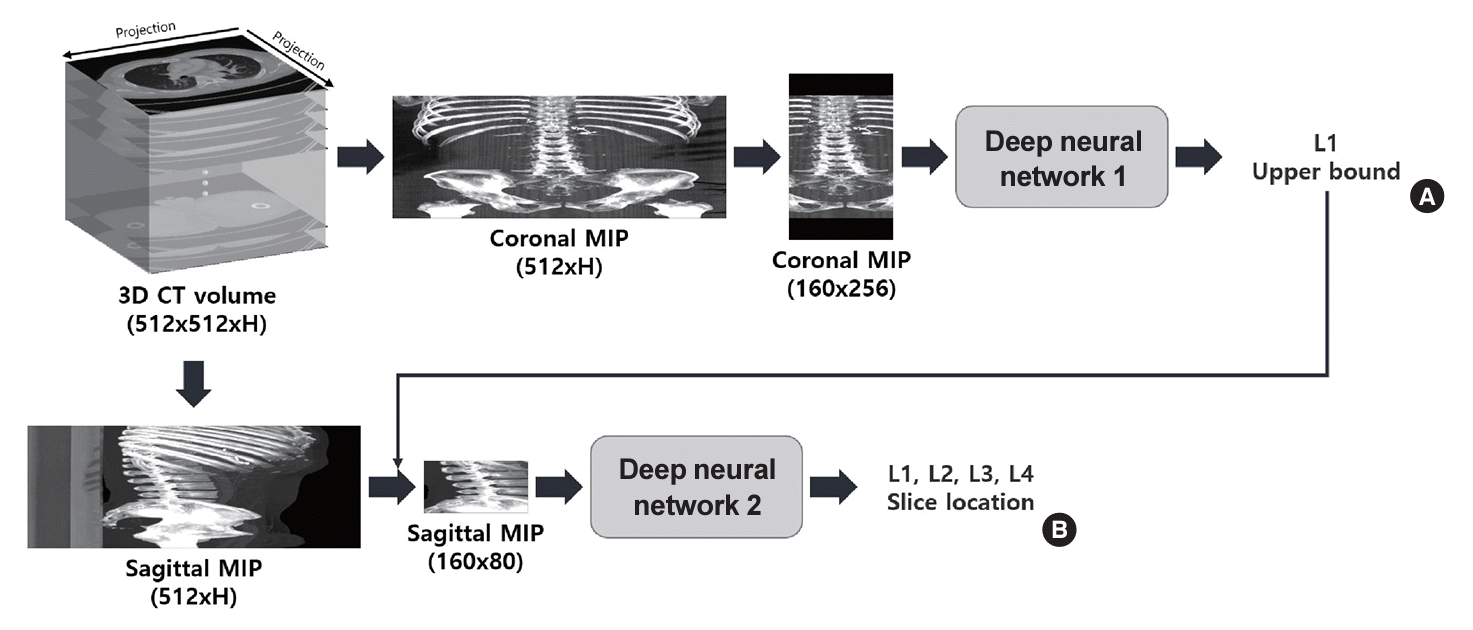
Fig. 3.
Training data for semi-supervised vertebra segmentation. For all subjects, the central axial computed tomography slice was annotated with segmentation slice labels. All axial slices for a given vertebra were used regardless of segmentation ground-truth availability during training.
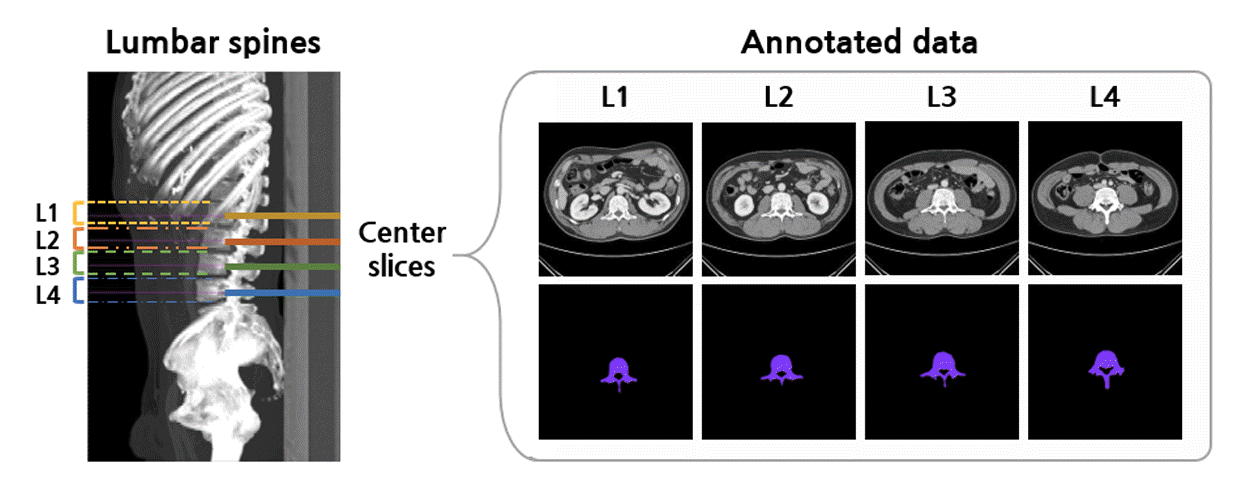
Fig. 4.
Data generation process for the bone mineral density estimation subtask. (A) Four computed tomography (CT) slices were selected from the detection subtask and their corresponding masks were generated from the segmentation subtask. (B) A 196×196 patch centered on the center of mass of the binary segmentation mask is cropped from each CT slice. (C) Cropped CT slices are concatenated counterclockwise to form a 392×392 input image.
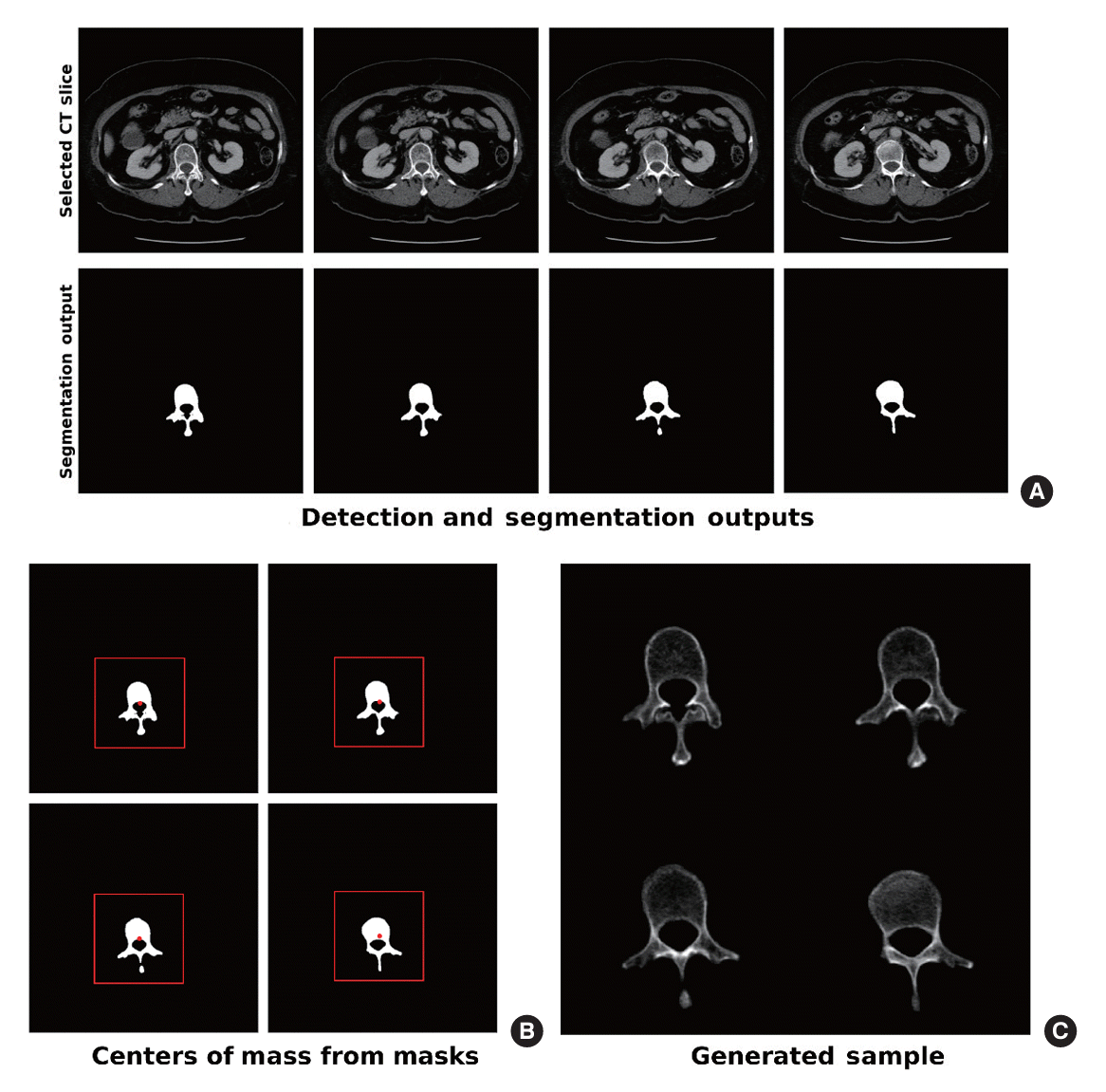
Fig. 5.
Regression (left) and Bland-Altman (right) plots for bone mineral density using dual-energy X-ray absorptiometry (BMDDXA) ground truths and end-to-end bone mineral density using convolutional neural network (BMDCNN) predictions. For both regression plots, the 95% confidence interval and 95% prediction interval are represented by the orange dotted line and grey dashed line respectively. (A) Vertebral-level results for L1, L2, L3, and L4 predictions evaluated independently (n=392). (B) Subject-level results for averaged L1 to L4 BMDCNN predictions against total lumbar BMDDXA (n=98). SD, standard deviation.
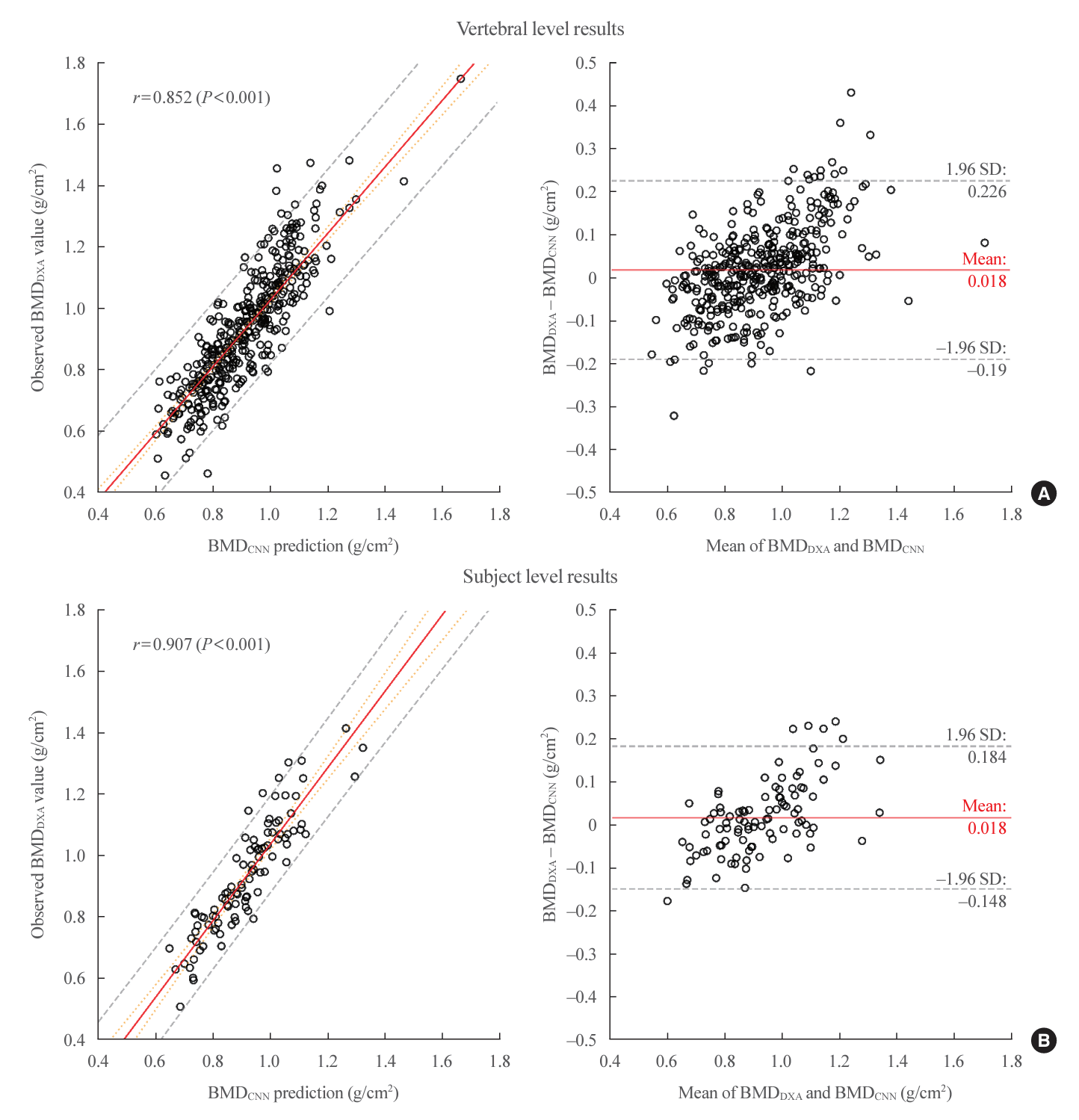
Table 1.
Demographic Characteristics
Table 2.
Spine Localization Results on the Held-out Test Set (n=98)
| Location | Mean | SD | Median | Max | ≥10a |
|---|---|---|---|---|---|
| L1 | 0.98 | 2.26 | 0 | 13 | 4 |
| L2 | 1.02 | 2.26 | 0 | 13 | 4 |
| L3 | 1.08 | 2.38 | 0.5 | 14 | 4 |
| L4 | 1.18 | 2.37 | 1 | 13 | 4 |
| Total | 1.07 | 2.31 | 1 | 14 | 16 |
Table 3.
Results of Segmentation Method Evaluated on Vertebral- and Subject-Levels
| Method |
Metric |
|
|---|---|---|
| Dice by vertebra | Dice by subject | |
| Baseline | 0.9673 | 0.9677 |
| Proposed | 0.9683 | 0.9685 |
Table 4.
Results of End-to-End BMDDXA Estimation across All Test Subjects (52 Females, 46 Males)
Vertebral-level results are based on end-to-end bone mineral density using convolutional neural network (BMDCNN) predictions evaluated independently from other vertebrae with ground-truth lumbar BMDDXA values. Subject-level results were generated by comparing total BMDDXA values with averaged L1, L2, L3, and L4 BMDCNN predictions for a given subject. All Pearson correlation coefficients had P values less than 0.001.
BMDDXA, bone mineral density using dual-energy X-ray absorptiometry; MAE, mean absolute error; RMSE, root mean squared error; MAPE, mean absolute percentage error.




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download