Abstract
Purpose
In this study, we investigated the frequencies of mutations in DNA damage repair genes including BRCA1, BRCA2, homologous recombination genes and TP53 gene in ovarian high-grade serous carcinoma, alongside those of germline and somatic BRCA mutations, with the aim of improving the identification of patients suitable for treatment with poly(ADP-ribose) polymerase inhibitors.
Materials and Methods
Tissue samples from 77 Korean patients with ovarian high-grade serous carcinoma were subjected to next-generation sequencing. Pathogenic alterations of 38 DNA damage repair genes and TP53 gene and their relationships with patient survival were examined. Additionally, we analyzed BRCA germline variants in blood samples from 47 of the patients for comparison.
Results
BRCA1, BRCA2, and TP53 mutations were detected in 28.6%, 5.2%, and 80.5% of the 77 patients, respectively. Alterations in RAD50, ATR, MSH6, MSH2, and FANCA were also identified. At least one mutation in a DNA damage repair gene was detected in 40.3% of patients (31/77). Germline and somatic BRCA mutations were found in 20 of 47 patients (42.6%), and four patients had only somatic mutations without germline mutations (8.5%, 4/47). Patients with DNA damage repair gene alterations with or without TP53 mutation, exhibited better disease-free survival than those with TP53 mutation alone.
Conclusion
DNA damage repair genes were mutated in 40.3% of patients with high-grade serous carcinoma, with somatic BRCA mutations in the absence of germline mutation in 8.5%. Somatic variant examination, along with germline testing of DNA damage repair genes, has potential to detect additional candidates for PARP inhibitor treatment.
Ovarian cancer is the leading cause of death among patients with gynecological malignancies worldwide, and approximately 225,000 ovarian cancers are newly diagnosed each year. The lack of an effective screening test and unfavorable anatomy are associated with advanced-stage disease at diagnosis and poor prognosis for the majority of patients. Overall survival is poor among gynecologic cancers, with a 5-year survival rate of 44% for all stages and 140,000 related deaths occurring annually worldwide, the majority of which are from high-grade serous carcinomas (HGSCs) [1-3].
Current estimates indicate that 20%-25% of women have an inherited germline mutation predisposing them to ovarian cancer [2]. In approximately 65%-85% of hereditary ovarian cancers, the associated genetic abnormality is a germline mutation of one of the BRCA genes (BRCA1 or BRCA2). Several other tumor suppressor genes and oncogenes are also associated with hereditary ovarian cancer, including homologous recombination (HR) DNA repair genes. The Cancer Genome Atlas (TCGA) reported HR deficiency in approximately 50% of patients with high-grade serous ovarian cancer [4]; however, in TCGA study, the majority of patients were European, with the percentage of Asian patients being only 3.2% (19/316).
The identification of a mutation in an ovarian cancer susceptibility gene represents a fundamental step in the diagnosis and treatment of these tumors. Moreover, the identification of a mutation in patients who have already been diagnosed can provide information about the pathogenesis of their tumors. With the development of targeted therapy, current strategies for the control and prevention of ovarian cancer rely on a thorough understanding of contributing genetic factors, at both the germline and somatic levels. In this context, next-generation sequencing technologies provide an unprecedented opportunity to simultaneously analyze multiple cancer susceptibility genes, reduce delays and costs, and optimize the molecular diagnosis of hereditary ovarian cancer.
Poly(ADP-ribose) polymerase (PARP) inhibitors have shown clinical effects in patients with ovarian cancer showing BRCA dysfunction or homologous recombination deficiency (HRD). PARP inhibitors were originally designed for synthetic lethal interaction with BRCA1 or BRCA2mutations [5,6]. In vitro studies have demonstrated that defects in the other HR proteins, such as ATM, CHEK1, CHEK2, NBN, and RAD51D, also confer sensitivity to PARP inhibitors [7,8]. Further, PARP inhibitors are active in a subset of sporadic (BRCA wild-type) recurrent platinum-sensitive ovarian carcinomas [9], which may be attributable to the influence of undetected HR gene alterations in that study. The possible application of PARP inhibitors as a therapeutic option for patients with ovarian cancer, and alterations in genes other than the BRCA genes, is currently under investigation (NCT-02476968, ORZORA study).
TP53 mutation is found in many cancer types and is related to DNA damage response and apoptosis [10]. It is well known that TP53 mutations are associated with poor prognosis in several cancers including ovarian cancers [10,11]. However, the relationship between DNA damage repair (DDR) gene and TP53 gene alterations and their combined effect on HGSC patient outcome has not been well described.
In this study, we investigated variants in DDR genes and TP53 gene in Korean patients with HGSC, analyzed their frequency and characteristics in relation to germline and somatic BRCA mutations in this group, and analyzed their impact on clinical outcome to provide better prediction for PARP inhibitor therapy response.
Eligibility criteria were as follows: women aged 20 years or older with pathological diagnosis of epithelial ovarian, fallopian tube, or peritoneal carcinoma, with a high-grade serous histologic component. Patients were treated using standard treatments (cyto-reductive surgery and/or platinum-based chemotherapy) at the time of diagnosis.
Family history of cancer was recorded and confirmed by direct contact with the patients and their families. A patient was considered to have a family history of cancer if any of the following criteria were met: (1) if there were one or more cases of ovarian, peritoneal, fallopian tube, breast, pancreas, or prostate cancer among first- or second-degree relatives; or (2) if the patient had a history of primary breast cancer.
Fresh frozen or formalin-fixed paraffin-embedded (FFPE) tumor tissue samples from the 77 patients with HGSC were analyzed. Among these 77 patients, blood samples were available from 47 patients for BRCA germline variant analysis. Fifty-nine cases with fresh tumor tissue, 48 available matched normal (pair in the same case) FFPE tissue for whole exome sequencing (diagnosed between the year 2005 and 2014), and 18 cases of FFPE tumor tissue for panel sequencing (diagnosed between 2017 and 2018) were obtained from the archive of Department of Pathology, CHA Bundang Medical Center. Two pathologists (H.K. and S.K.) reviewed the histology (Fig. 1) using the 2014 World Health Organization classification criteria [1].
Genomic DNA was extracted from a single surgical sample, containing a HGSC component, from each patient. Two different targeted sequencing assays were carried out: (1) whole exome sequencing and (2) targeted gene panel sequencing.
Fifty-nine fresh tumor samples for whole exome sequencing were snap frozen, and genomic DNA was extracted using GeneAll Exgene Clinic SV Kit columns (GeneAll, Cambio, UK), according to the manufacturer’s protocols. Exomes were captured using the Agilent SureSelect Exome V5 probe set, and the captured DNA was sequ-enced on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA). Sequences were aligned using BWA-0.7.15, based on Genome Reference Consortium build 37 (GRCh37). Duplicated reads were removed using Picard-tools 2.7.1, and variants were called, according to GATK 3.8 best practices (https://software.broadinstitute.org/gatk/best-practices/). Among the 59 patients, we performed whole exome sequencing with the matched normal FFPE lymph node tissue of 48 patients for whom the matched normal tissue was available.
For 18 FFPE tumor tissue samples for panel sequencing, genomic DNA was extracted using the RecoverAll multisample RNA/DNA isolation workflow (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocols. One hundred forty-three genes were targeted using the Ion Torrent Oncomine Comprehensive Assay v1 (OCA1, Thermo Fisher Scientific, Waltham, MA) and sequenced using an Ion S5 XL. Sequences were aligned using Torrent Suite 5.10.0, based on GRCh37, and variants were called with Torrent Variant Caller 5.10-9.
Thirty-eight DDR genes (including BRCA1/2, HR genes, mismatch repair genes) and TP53 gene were selected for analysis of the pathogenic alterations with fresh or FFPE tissues in 77 patients.
Among them, for comparison of germline and somatic BRCA1 and BRCA2 variants of 29 whole exome sequencing patients and 18 panel sequencing patients were analyzed. Germline Sanger sequencing was separately conducted with blood samples in 47 patients among 77 patients (Tables 1 and 2).
For identifying pathogenic variants from germline common variants and reducing false-positive variants without available matched normal samples, we took three strategies of filtering out common germline or false-positive variants. Firstly, we removed variants with low qualities (QUAL score < 30 or depth < 20 or FILTER=FAIL). Secondly, we removed common variants (allele frequency > 0.01) in dbSNP database (00-common_all.vcf file was downloaded on June 7, 2018) or in ExAC database (ExAC.r1.sites.vep.vcf file was downloaded on July 16, 2018). Lastly, we removed common variants in Korean (allele frequency > 0.01) based on KOVA (K1055E_allele_frequency.txt was downloaded on November 15, 2017) and KRGDB (variants1100_cmm.txt file was downloaded on December 1, 2016 from http://coda.nih.go.kr/coda/KRGDB).
Fresh tumor somatic variant calls and the matched normal germline variant calls for 48 whole exome sequencing patients were carried out using the same GATK 3.8 best practice pipeline and GATK4.1-Mutect2 Tumor-Normal pair pipeline (FILTER=PASS and Depth ≥ 20), respectively. The information of germline or somatic variants is provided in S1 and S2 Tables.
For functional annotation of each variant, ANNOVAR [12], Variant Effect Predictor [13], and Ion Reporter ver. 5.2 software (Thermo Fisher Scientific) were used. For interpretation of the pathogenicity of each variant, the three knowledge database were used; ClinVar, Oncomine Knowledge-base, and guidelines for the interpretation of sequence variants in cancer (American College of Medical Genetics) referred to Richards et al.’s study [14]. Among the whole exome sequencing and panel sequencing test results, information on 38 DDR genes and TP53 gene was obtained (S1-S3 Tables). These 39 genes were selected for inclusion for a more focused analysis on BRCA and HRD genes. To assess the clinical impact of these genes, two subgroups according to the mutation status of TP53 and the other 38 DDR genes of patients were separately analyzed for their survival. The groups were defined as follows: group 1 includes patients with pathogenic mutation in any of the 38 DDR genes, regardless of TP53 mutation (n=31), and group 2 includes patients with TP53 mutation only (n=37).
Of the 77 patients who underwent somatic genetic analysis, 47 had previously undergone germline BRCA testing, and genetic information on germline BRCA status was obtained and compared with the somatic BRCA test results. All germline tests were carried out using the Sanger sequencing method, as previously described [15].
To evaluate the differences in overall survival and diseasefree survival of the group 1 (any DDR gene mutation–positive regardless of TP53 mutation status) and group 2 (TP53 mutation only) were statistically significant, the log-rank test was used. Survival analysis was conducted using the survival and survminer R packages and the p-value of each Kaplan Meier-plot was calculated by log-rank test. Survival plots were depicted in ggplot2 R package.
Women who met the criteria (n=77) provided written informed consent between 2009 and 2018, and the study was conducted with approval from the Institutional Review Board of the CHA Bundang Medical Center (IRB approval No. 2016-10-010-005). Use of patient-derived samples for this study was approved, and the work described was performed in accordance with approved guidelines.
The clinical characteristics of the 77 patients included in the study are listed in Table 3. The majority ethnicity was Korean, with only four individuals of Korean Chinese ethnicity. Median age at diagnosis was 56 years (range, 36 to 82 years). Primary cancers were ovary (92.2%), fallopian tube (3.9%), and peritoneum (3.9%). Histologically, the majority of tumors were HGSC. One of two patients with carcinosarcoma had HGSC in the left ovary and metastatic lesions and carcinosarcoma in only the right ovary. In this patient, only the HGSC component was used for tumor genetic testing. Approximately 90% of patients had advanced-stage disease (> stage III). Six patients (7.8%) were previously diagnosed with breast cancer; 17 (22.1%) had a family history of breast, ovarian, fallopian tube, peritoneal, pancreas, or prostate cancer in first- or second-degree relatives. Twenty-one patients (21/77, 27.3%) had positive “family history,” according to the definition used in the present study.
One or more mutation in the DDR genes was identified in 40.3% of patients (31 of 77). Of the 38 DDR genes and TP53 gene, pathogenic mutations were identified in eight genes; the remaining 31 genes were wild type. Specific details of the 98 pathogenic mutations and 63 variants of unknown significance identified in these 39 genes in the 77 patients are listed in S1-S3 Tables. Observed pathogenic and likely pathogenic mutations are shown in Fig. 2. BRCA1 (28.6%), BRCA2 (5.2%), and TP53 (80.5%) were the most frequently altered genes. BRCA and TP53 mutations were detected pervasively. The frequencies of the detected variants are shown in S4 Fig. Among BRCA gene mutations, BRCA1 Y130Ter variants were observed in three patients (13.6%, 3/22). The TP53 alterations observed with frequency of more than two patients are as follows. TP53 R175H (5.6%) and TP53 R248Q/W (7.9%) were observed in three (4.8%) and two (3.2%) patients among 62 patients, respectively. TP53 K132R, I195T, Y220C, and R306* variants were observed in three patients (4.8%). TP53 mutations were significantly enriched in DNA-binding domain (binomial test p-value=1.38e-4). The details of the TP53 gene mutation in this study is shown in graphic chart in S5 Fig.
DDR gene alterations other than BRCA1/2 genes were detected in (in order of frequency) RAD50 (5.1%), ATR (3.4%), MSH2 (2.6%), FANCA (1.7%), and MSH6 (1.7%). One patient (Pat53) carried four mutations (TP53, RAD50, ATR, and MSH2) simultaneously, while another two patients each had three different mutations.
To assess the clinical impact of these genes, two subgroups of patients were analyzed for their survival (Fig. 3A), according to the mutation status of 38 DDR genes and TP53 gene as follows: group 1 (pathogenic mutation in any of the 38 DDR genes, regardless of TP53 mutation); group 2 (TP53 mutation only). The overall survival of patients in these two groups did not differ significantly (p=0.12) (Fig. 3B); however, disease-free survival was different (p=0.05) (Fig. 3C). The disease-free survival of patients with at least one pathogenic mutation in any of the 38 DDR genes was better than that of patients with TP53 mutations only.
Nine patients were interpreted negative for DDR genes and TP53 gene mutation. They were not included for survival analysis because the number of patients was too small for meaningful statistical analysis.
Pathogenic variants of BRCA genes were detected in 20 of the 47 patients (42.6%) in total: 17 in BRCA1 and 3 in BRCA2. Among these, germline variants were identified in 16 (34.0%) of the 47 patients; 14 of the variants were in BRCA1, with two in BRCA2. Of note, four patients (8.5%) had only somatic BRCA variations without germline mutation (Table 4).
The most common histologic type of ovarian cancer is epithelial ovarian carcinoma (EOC), which has five subtypes: high-grade serous (70%), endometrioid (10%), clear cell (10%), low-grade serous (5%), and mucinous (3%). EOC is both clinically diverse and molecularly heterogeneous, and its subtypes have distinct gene expression patterns. Ovarian HGSC is distinct from non-HGSC, differing with respect to clinical presentation, disease distribution, response to therapy, survival, and site of origin [16]. Pennington et al. [17] repor-ted that non-HGSC cases had some BRCA mutations and a greater proportion of mutations in other HR genes. Therefore, we focused on tumors with HGSC histology and excluded non-HGSC tumors from the present study.
The frequency of somatic TP53 mutations in Korean patients with HGSC in the present study was lower (80.5%) than expected. High-grade serous ovarian carcinoma is characterized by ubiquitous TP53 abnormalities, with BRCA changes in approximately 50% of cases, along with chromosomal instability. TP53 aberration has been considered a driver event in ovarian carcinogenesis, and it is possible that TP53 mutations are more stable over time than other mutations. TP53 was mutated in 95.9% of HGSC samples (303 of 316) in TCGA 2011 study [4]; however, in the present study, the frequency of TP53 mutation among HGSC patients was 80.5%. When we investigated the frequency of TP53 mutations in 10 Asian patients included in TCGA 2011 study, nine (90%) of them had TP53 mutations. There was no statistically significant association between incidence of TP53 mutations and ethnicity (Asian vs. others [mostly European]) in TCGA data set (p=0.35). Regarding the quality of sequencing, OCA1 panel that we used in this study includes the whole exon regions of TP53 as well as whole exome sequencing, and the average coverage of all targeted regions of TP53 in two sequencing methods were 98X for whole exome sample, and 3576X for OCA1 panel sample. We provided the sequencing coverage of overall targets and of TP53 in each patient in S3 Table. We also confirmed that there was no statistical difference of sequencing coverage between patients with TP53 mutation versus without TP53 mutations. It is shown in S6 Fig. There was no statistical difference of sequencing coverage between patients with TP53 mutations versus without TP53 mutations, even though the average coverage values in two sequencing methods are different (S6 Fig.). However, when we investigated the TP53 variants with low quality (depth < 20), there were TP53 mutations in seven patients. If we included seven TP53 mutations with low quality, the frequency of TP53 mutations would be 89.6%, comparable to TCGA 2011 study. The information on mutations with low quality (depth < 20) are provided in S7 Table.
We compared the disease-free survival of the subset of HGSCs with DDR gene mutation with or without TP53 mutation (group 1) and TP53 mutations only (group 2) to examine the effect of these alterations on disease-free survival of the HGSC patients. The disease-free survival of group 1 was better than that of group 2 (Fig. 3C), which is in accordance with previous studies that BRCA1, BRCA2 gene mutation and HRD are related to favorable clinical outcome [18,19]. TP53 mutation status and its impact on survival is different between low grade and HGSCs, but among the HGSCs, BRCA and HR genes have more impact on survival than TP53 mutation status.
When DDR genes are affected, the cancer cells may become more sensitive to anticancer agents directed to DNA damage such as platinum-based antineoplastic therapy. Because many anticancer therapy agents are directed to DNA damage such as alkylating agents. If the DNA damage increases without repair, it may lead to cell death. Such tumors may also produce more neoantigens due to increased mutational load. It may also elicit more immune responses to tumor and these patients may also be the candidates for cancer immunotherapy [11].
Estimates of the contribution of germline BRCAmutations to EOC vary widely, from 5% to 20% [20], and patients having somatic mutations without germline mutations in BRCA are less frequent, accounting for 2%-8% [21-24]. Germline and somatic BRCA mutation frequencies in EOC patients, according to the published literature, are listed in Table 5 [4,23-27]. As for HGSC, the lowest frequency of germline and somatic BRCA mutations (21.8%) is recorded in TCGA 2011 data, with the highest (20/47, 42.6%) in the present study. The mean ratio of HGSC patients with only somatic BRCA mutations to both germline and somatic mutations in the literature is 24.8% (range, 16.1% to 33.3%). Germline BRCA testing is widespread in recent clinical practice; however, this approach could miss the patients with only somatic BRCA mutations, who could also potentially benefit from therapy with PARP inhibitors.
Although previous reports on the effectiveness of PARP inhibitors in cancers with BRCA and HR repair defects have mainly focused on germline alteration of these genes, such treatment may also be effective for patients with somatic aberrations. Some patients with ovarian cancer who do not carry germline BRCA mutations also respond to PARP inhibitors [28], suggesting that broader dysfunction of genes, other than the BRCA genes. The patients with DDR mutation other than BRCA mutation may also be responsive to PARP inhibitors. The high frequency of HRD in HGSC in this study suggests that screening for this phenotype should be considered to identify candidates for PARP inhibitor treatment among these patients.
In the present study, HGSC showed somewhat higher germline and/or somatic BRCAmutation frequency in Korea (20/47 cases, 42.6%), compared with the results of previous studies (21.8%-30.1%) (Table 5). When we reviewed our germline and somatic genetic test results, and there were no false-positive results or variants of unknown significance. In the present study, the germline BRCA mutation rate (16/47 cases, 34.0%) was higher than previous reports in other countries (15.8%-25.2%), but the somatic BRCA mutation rate (4/47 cases, 8.5%) did not show much difference with the previous reports (4.9%-8.7%). It was reported that germline BRCA mutation frequency of Korean epithelial ovarian cancer patients was higher than previous reports [15]. In another recent study of ovarian advanced HGSC in Korean patients, germline BRCAmutation was found in 39.8% (51/128 cases), which is similar to the present study [29]. The possible reasons for higher rate of germline BRCAmutation in this study may be due to ethnic variation. Another reasons for differences in germline BRCA mutation rate maybe inclusion of primary fallopian tube or primary peritoneal HGSC cases which seem to show higher rate of BRCA mutation rate than primary ovarian cancer, or heterogeneity of cases included in different studies. Whether the study includes only HGSC or other epithelial ovarian cancer, inclusion of fallopian tube or primary peritoneal cancer, proportion of patients with family history may influence the detection rate of germline BRCA mutation.
We did not perform multiplex ligation-dependent probe amplification (MLPA) test to detect large genomic rearrangements (LGRs) in this study. Prevalence of LGRs was reported to be 1.8% in Korea at recent study [30]. The frequency of BRCA mutation would have been slightly higher if the MLPA test had been performed.
The reported prevalence of germline BRCA mutations in patients with fallopian tube or peritoneal cancers ranges from 15.8% to 40.9% [31]; however, BRCA mutations (two germline and two somatic) were found in four of six patients (66.7%) with fallopian tube or peritoneal cancers in the present study (Table 4). The one remaining patient (Pat17) with fallopian tube cancer had somatic TP53 and FANCA mutations (S3 Table). Although a small number of subjects with peritoneal and fallopian tube cancers were included in this study, these cancers exhibited higher frequency of BRCA mutation than ovarian cancers, and all of the cases had at least one alteration among the 39 genes included in this study. These data require confirmation by further study using a larger number of cases; however, it may imply that more candidates for PARP inhibitor therapy may be detected among patients with fallopian tube or peritoneal HGSC.
In conclusion, in this study we examined variants in DDR genes and TP53 gene in patients with ovarian HGSC. To the best of our knowledge, this study is the first to analyze somatic BRCA1, 2 and DDR gene mutations and compare with germline and somatic BRCA mutations in Korean patients with HGSC. Mutations of DDR genes were observed in 40.3% (31/77), including BRCAmutations, and the frequency of TP53 mutation (80.5%) was low compared with that in previous reports. Patients carrying somatic BRCAmutations without germline mutations were identified at a frequency of 8.5%. Although further validation in a large-scale study is needed, our data strongly indicate that more candidates for PARP inhibitor treatment can be detected by examination of somatic BRCA gene variants and other DDR genes, alongside germline testing for BRCA genes.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
S1 Table.
Ninety-eight pathogenic and likely pathogenic variants of 39 genes in 77 patients
S4 Fig.
The distribution of altered codon in BRCA1 and TP53 gene without hotspot (A) The bar graph shows the number of pathogenic or likely pathogenic variants in each TP53 codon. TP53 R175H (5.6%) and TP53 R248Q/W (7.9%) were observed in three (4.8%) and two (3.2%) cases among 62 patients, respectively. TP53 K132R, I195T, Y220C, and R306* variants were observed in three patients (4.8%). (B) The bar graph shows the number of pathogenic or likely pathogenic variants in each BRCA1 codon. BRCA1 Y130Ter variants were observed in three patients (3/22, 13.6%).
S5 Fig.
The details of the TP53 gene mutation in this study is shown in graphic chart. TP53 mutations were significantly enriched in DNA-binding domain.
S6 Fig.
The TP53 sequencing coverage in the two sequencing methods of panel sequencing and exome sequencing. (A) In panel sequencing, there was no statistical difference of TP53 sequencing coverage between patients with TP53 mutations and without it (t-test p-value=0.58). (B) In whole exome sequencing, there was also no statistical difference between patients with TP53 mutations and without it. (t-test p-value=0.48).
ACKNOWLEDGMENTS
This work was supported by a grant (HI16C1559) from the Korea Health Technology R&D Project through The Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, and supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1C1B5032617), Republic of Korea.
References
1. Kurman RJ, Carcangiu ML, Herrington CS, Young RH. WHO classification of tumours of female reproductive organs. 4th ed. Lyon: International Agency for Research on Cancer;2014.
2. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011; 108:18032–7.

4. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011; 474:609–15.
5. Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010; 28:2512–9.
6. Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014; 15:852–61.

7. Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011; 43:879–82.

8. McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006; 66:8109–15.

9. Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011; 12:852–61.

10. Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007; 26:2157–65.

11. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018; 23:239–54.
12. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.

13. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016; 17:122.

14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.

15. Choi MC, Heo JH, Jang JH, Jung SG, Park H, Joo WD, et al. Germline mutations of BRCA1 and BRCA2 in Korean ovarian cancer patients: finding founder mutations. Int J Gynecol Cancer. 2015; 25:1386–91.
16. Przybycin CG, Kurman RJ, Ronnett BM, Shih IM, Vang R. Are all pelvic (nonuterine) serous carcinomas of tubal origin? Am J Surg Pathol. 2010; 34:1407–16.

17. Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014; 20:764–75.

18. Safra T, Lai WC, Borgato L, Nicoletto MO, Berman T, Reich E, et al. BRCA mutations and outcome in epithelial ovarian cancer (EOC): experience in ethnically diverse groups. Ann Oncol. 2013; 24 Suppl 8:viii63–8.

19. Patel JN, Braicu I, Timms KM, Solimeno C, Tshiaba P, Reid J, et al. Characterisation of homologous recombination deficiency in paired primary and recurrent high-grade serous ovarian cancer. Br J Cancer. 2018; 119:1060–6.

20. Ramus SJ, Gayther SA. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol Oncol. 2009; 3:138–50.
21. Foster KA, Harrington P, Kerr J, Russell P, DiCioccio RA, Scott IV, et al. Somatic and germline mutations of the BRCA2 gene in sporadic ovarian cancer. Cancer Res. 1996; 56:3622–5.
22. Berchuck A, Heron KA, Carney ME, Lancaster JM, Fraser EG, Vinson VL, et al. Frequency of germline and somatic BRCA1 mutations in ovarian cancer. Clin Cancer Res. 1998; 4:2433–7.
23. Mafficini A, Simbolo M, Parisi A, Rusev B, Luchini C, Cataldo I, et al. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget. 2016; 7:1076–83.

24. Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD 2nd, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010; 28:3570–6.

25. Zhao Q, Yang J, Li L, Cao D, Yu M, Shen K, et al. Germline and somatic mutations in homologous recombination genes among Chinese ovarian cancer patients detected using next-generation sequencing. J Gynecol Oncol. 2017; 28:e39.

26. McAlpine JN, Porter H, Kobel M, Nelson BH, Prentice LM, Kalloger SE, et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol. 2012; 25:740–50.

27. Chao A, Chang TC, Lapke N, Jung SM, Chi P, Chen CH, et al. Prevalence and clinical significance of BRCA1/2 germline and somatic mutations in Taiwanese patients with ovarian cancer. Oncotarget. 2016; 7:85529–41.

28. Tinker AV, Gelmon K. The role of PARP inhibitors in the treatment of ovarian carcinomas. Curr Pharm Des. 2012; 18:3770–4.

29. Kim SI, Lee M, Kim HS, Chung HH, Kim JW, Park NH, et al. Effect of BRCA mutational status on survival outcome in advanced-stage high-grade serous ovarian cancer. J Ovarian Res. 2019; 12:40.

Fig. 1.
Histologic features of high-grade ovarian serous carcinoma. (A) High-grade serous carcinoma (HGSC) composed of papillary, glandular patterns with large, hyperchromatic, pleomorphic nuclei and numerous mitoses is shown (H&E staining, ×200). (B) Diffuse strong positive reaction to immunohistochemical stain for p53 in HGSC is shown (p53 immunohistochemical stain, ×200).
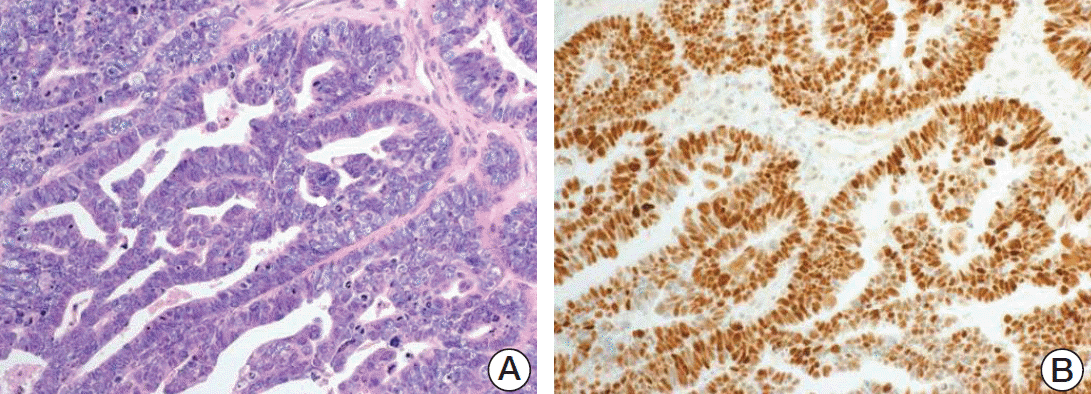
Fig. 2.
Pathogenic variants (n=98) in 77 ovarian cancer patients. TP53 was mutated in 80.5% of cases, the highest frequency among the 39 genes analyzed. BRCA1 and BRCA2 were mutated in 28.6% and 5.2% of cases, respectively. The color of each cell represents the type of variant: blue, indel; red, stop-gain; green, missense; orange, splicing; gray, NA (data not available); and white, wild type.
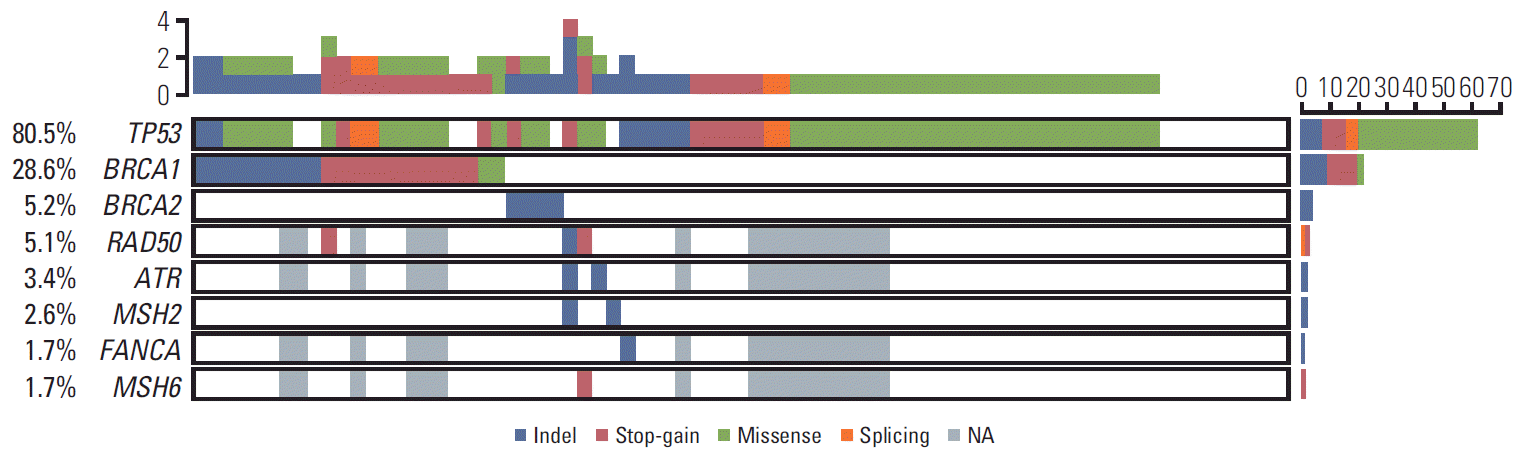
Fig. 3.
The prognosis of two groups of patients, classified according to pathogenic variants. (A) Two groups were defined by their status for the pathogenic variants described in Fig. 1. Group 1 consisted of patients with any pathogenic mutations in the 38 DNA damage repair genes regardless of TP53 mutation status. Group 2 consists of patients with only TP53 mutations. Overall survival (OS) and disease-free survival (DFS) of the two groups are shown in B and C, respectively. NA, not available.
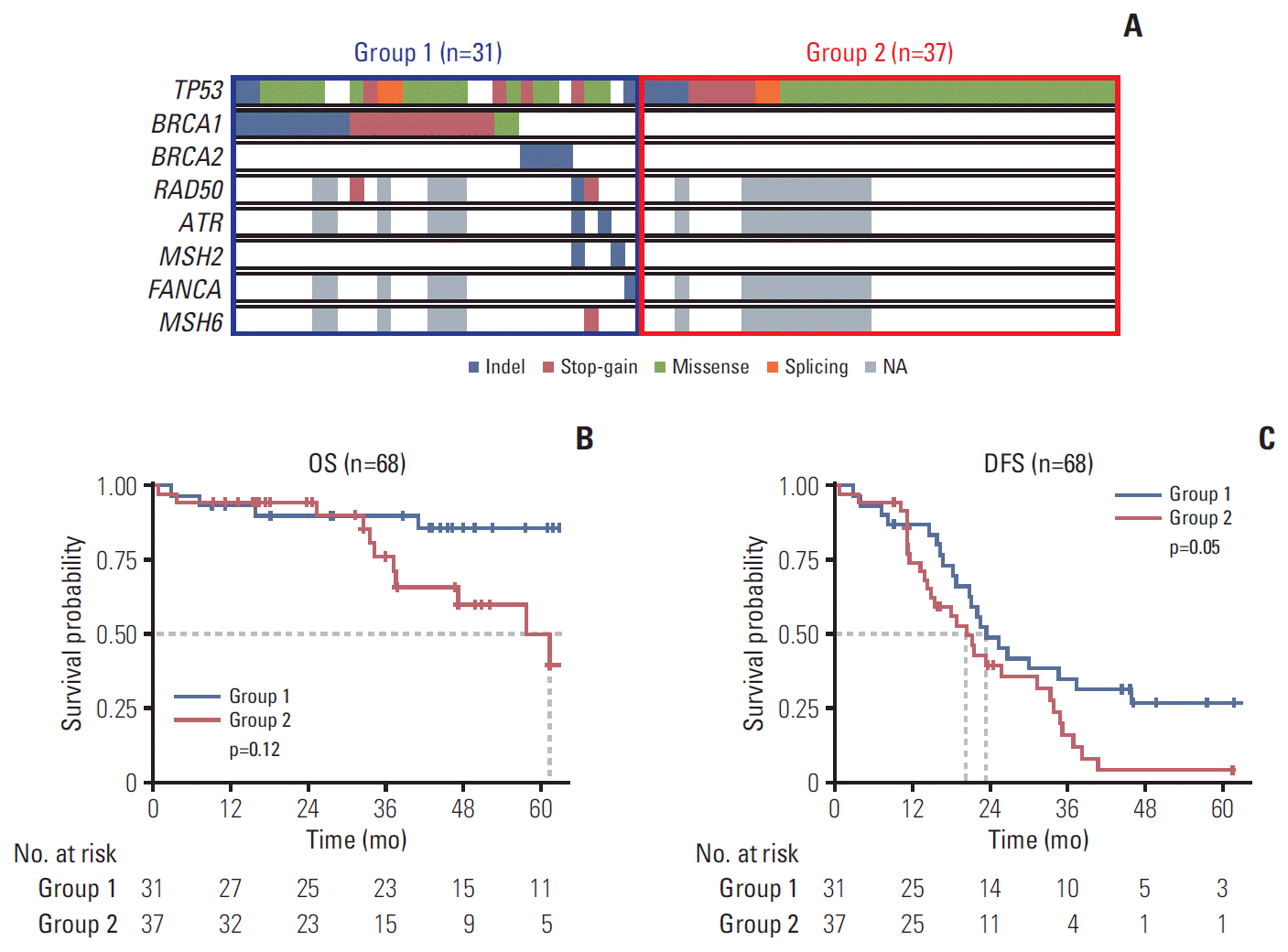
Table 1.
Thirty-nine genes associated with hereditary ovarian cancer
Table 2.
Samples and sequencing methods
Table 3.
Patient clinical characteristics
| Characteristic | No. (%) (n=77) |
|---|---|
| Ethnicity | |
| Korean | 73 (94.8) |
| Korean-Chinese | 4 (5.2) |
| Age, median (range, yr) | 56 (36-82) |
| Cancer site | |
| Ovary | 71 (92.2) |
| Fallopian tube | 3 (3.9) |
| Peritoneum | 3 (3.9) |
| Histology | |
| High-grade serous | 73 (94.8) |
| Seromucinous | 2 (2.6) |
| Carcinosarcoma | 2 (2.6) |
| Stage | |
| I | 2 (2.6) |
| II | 6 (7.8) |
| III | 59 (76.6) |
| IV | 10 (13.0) |
| Breast cancer history | |
| Yes | 6 (7.8) |
| No | 71 (92.2) |
| Family history of BRCA-related cancera) | |
| Yes | 17 (22.1) |
| No | 60 (77.9) |
| Residual mass (cm) | |
| < 1 | 67 (87.0) |
| ≥ 1 | 10 (13.0) |
| Platinum sensitivity | |
| Sensitive | 40 (64.5) |
| Resistant | 16 (25.8) |
| Lost to follow-up | 6 (9.7) |
| Status | |
| NED | 15 (19.5) |
| AWD | 35 (45.5) |
| Death | 21 (27.3) |
| Lost to follow-up | 6 (7.8) |
Table 4.
Detected BRCA1/2 mutations in 47 patients by germline and somatic genetic test
| Case | Age (yr) | Cancer | Stage |
Detected in blood and/or matched normal tissue |
Detected in tumor |
Germline/Somatic | Family historya) | ||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Mutation | Gene | Mutation | ||||||
| OCA07 | 53 | Ovary | III | BRCA1 | c.2048delA | BRCA1 | c.2048delA | Germline | + |
| Pat55 | 59 | Ovary | III | BRCA1 | c.2359delG | BRCA1 | c.2359delG | Germline | – |
| OCA02 | 47 | Ovary | III | BRCA1 | c.928C>T | BRCA1 | c.928C>T | Germline | – |
| Pat10 | 53 | Ovary | III | BRCA1 | c.1716dupA | BRCA1 | c.1716dupA | Germline | + |
| Pat48 | 64 | Ovary | III | BRCA1 | c.3442G>T | BRCA1 | c.3442G>T | Germline | + |
| Pat20 | 52 | Ovary | III | BRCA1 | c.3700_3704delGTAAA | BRCA1 | c.3700_3704delGTAAA | Germline | + |
| Pat45 | 61 | Ovary | III | BRCA1 | c.922_924delAGCinsT | BRCA1 | c.922_924delAGCinsT | Germline | + |
| Pat21 | 55 | Ovary | III | BRCA1 | c.4801A>T | BRCA1 | c.4801A>T | Germline | + |
| Pat46 | 47 | Ovary | III | BRCA1 | c.390C>A | BRCA1 | c.390C>A | Germline | + |
| Pat11 | 65 | Ovary | III | BRCA1 | c.390C>A | BRCA1 | c.390C>A | Germline | + |
| Pat05 | 77 | Ovary | IV | BRCA2 | c.3860delA | BRCA2 | c.3860delA | Germline | + |
| Pat08 | 55 | Ovary | III | BRCA2 | c.5576_5579delTTAA | BRCA2 | c.5576_5579delTTAA | Germline | + |
| OCA18 | 57 | FTC | III | BRCA1 | c.3895C>T | BRCA1 | c.3895C>T | Germline | + |
| OCA13 | 43 | PPC | III | BRCA1 | c.3813dupT | BRCA1 | c.3813dupT | Germline | – |
| Pat41 | 58 | Ovary | III | BRCA1 | c.5496_5506delinsA | BRCA1 | c.5496_5506delinsA | Germline | + |
| Pat49 | 48 | Ovary | III | BRCA1 | c.390C>A | BRCA1 | c.390C>A | Germline | – |
| Pat47 | 49 | Ovary | III | Not detected | BRCA1 | c.4287C>A | Somatic | – | |
| OCA16 | 55 | Ovary | IV | Not detected | BRCA1 | c.969_970delAA | Somatic | – | |
| OCA19 | 52 | FTC | III | Not detected | BRCA1 | c.1813delG | Somatic | + | |
| OCA14 | 50 | PPC | III | Not detected | BRCA2 | c.547delA | Somatic | + | |
Table 5.
Frequency of BRCA germline and somatic mutations in ovarian cancer reported in the literature
| Cancer site | Histology | Nation of publication | Germline mutation | Somatic mutation | S/(S+G) ratio | G and S mutation | Study |
|---|---|---|---|---|---|---|---|
| Ovary | Serous and non-serous | USA | 17/28 (60.7) | 11/28 (39.3) | 11/28 (39.3) | 28/28 (100) | Hennessy et al. (2010) [24] |
| Ovary | Serous and non-serous | China | 12/50 (24.0) | 2/50 (4.0) | 2/14 (14.3) | 14/50 (28.0) | Zhaoe et al. (2017) [25] |
| Ovary | HGS | USA | 50/316 (15.8) | 20/316 (6.3) | 20/69b) (29.0) | 69/316 (21.8) | TCGA (2011) |
| POFT | HGS | Canada | 26/103 (25.2) | 5/103 (4.9) | 5/31 (16.1) | 31/103 (30.1) | McAlpine et al. (2012) [26] |
| Ovary | HGS | Italy | 10/47 (21.3) | 3/47 (6.4) | 3/13 (23.1) | 13/47 (27.7) | Mafficini et al. (2016) [23] |
| Ovary | HGS | Taiwan | 8/46 (17.4) | 4/46 (8.7) | 4/12 (33.3) | 12/46 (26.1) | Chao et al. (2016) [29] |
| POFT | HGS | Korea | 16/47 (34.0) | 4/47 (8.5) | 4/20 (20.0) | 20/47 (42.6) | Present study (2019) |
| Totala) | 110/559 (19.7) | 36/559 (6.4) | 36/145 (24.8) | 145/559 (25.9) |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download