Abstract
Background
Metabolic dysregulation is a hallmark of type 2 diabetes mellitus (T2DM), in which the abnormalities in brown adipose tissue (BAT) play important roles. However, the cellular composition and function of BAT as well as its pathological significance in diabetes remain incompletely understood. Our objective is to delineate the single-cell landscape of BAT-derived stromal vascular fraction (SVF) and their characteristic alterations in T2DM rats.
Methods
T2DM was induced in rats by intraperitoneal injection of low-dose streptozotocin and high-fat diet feeding. Single-cell mRNA sequencing was then performed on BAT samples and compared to normal rats to characterize changes in T2DM rats. Subsequently, the importance of key cell subsets in T2DM was elucidated using various functional studies.
Results
Almost all cell types in the BAT-derived SVF of T2DM rats exhibited enhanced inflammatory responses, increased angiogenesis, and disordered glucose and lipid metabolism. The multidirectional differentiation potential of adipose tissue-derived stem cells was also reduced. Moreover, macrophages played a pivotal role in intercellular crosstalk of BAT-derived SVF. A novel Rarres2+macrophage subset promoted the differentiation and metabolic function of brown adipocytes via adipose-immune crosstalk.
· BAT-SVF cells showed increased inflammation, angiogenesis, and abnormal metabolism in T2DM.
· Differentiation potential is reduce in adipose-derived stem cells of T2DM rats.
· Macrophages play a crucial role in the intracellular crosstalk of BAT SVF.
· Rarres2+ macrophages enhance brown adipocyte function via adipose-immune crosstalk.
Adipose tissues, crucial in type 2 diabetes mellitus (T2DM) metabolic disorders, undergo significant changes in their cellular composition and function during the development of T2DM and its complications [1]. Adipose tissues include white adipose tissue (WAT), brown adipose tissue (BAT), and beige adipose tissue. However, most T2DM studies have focused on WAT.
Active BAT, abundant in adult’s neck and mediastinum, consists of brown adipocytes and the stromal vascular fraction (SVF). BAT enhances glucose uptake and insulin sensitivity, promotes fatty acid oxidation, and reduces blood glucose and lipid levels [2]. It also produces active molecules that regulate the function of other tissues [3]. In diabetes, BAT is reported to differentiate into WAT, and its uptake and utilization of glucose are significantly reduced [4]. BAT transplantation can improve glucose metabolism and insulin sensitivity, and reduce body mass and adiposity [5]. However, a study also showed that removing BAT from mice did not affect glucose metabolism [6].
Macrophages (MACs), the most abundant immune cells in adipose tissue, are crucial in diabetes development [7]. MACs reportedly increase BAT’s nonshivering thermogenesis and promote WAT browning by secreting norepinephrine, but some studies dispute this [8,9]. MACs are typically classified into proinflammatory M1 and anti-inflammatory M2 phenotypes [10]. In obesity or T2DM patients, M2 MACs in WAT are polarized to the M1 phenotype, increasing inflammation and insulin resistance [11]. However, recent research suggests the complexity of MAC types and functions exceeds the M1/M2 classification [12]. Moreover, current research on MACs in adipose tissue mainly focuses on WAT, with few studies on MACs in BAT.
Single-cell RNA sequencing (scRNA-seq) is a powerful tool for identifying new cell types and functions. It has been used to identify a new cell population in WAT SVF that inhibits white adipocyte differentiation, and the platelet derived growth factor receptor alpha (PDGFRα)+stem cell antigen-1 (Sca1)+cluster of differentiation 81 (CD81)+adipose stem/progenitor cells (ASPCs) with beige fat plasticity [13]. It has also revealed galectin 3 (Lgals)+cluster of differentiation 36 (CD36)+age-dependent regulatory cells in WAT that express adipose progenitor markers but suppress the proliferation and differentiation of adipose precursor cells, and the low- and high-thermogenic brown adipocyte subpopulations in murine adipose tissue [14,15]. However, the cell types and functions of the BAT SVF and their abnormalities in T2DM remain largely unknown.
In the present study, we revealed the single-cell landscape of BAT SVF and its characteristic changes in T2DM rats using scRNA-seq. We also identified a retinoic acid receptor responder 2 (Rarres2)+MAC subset that promoted brown adipocyte differentiation and function. These findings deepen the understanding of BAT function and its role in T2DM metabolic disorders.
Four-week-old male Wistar rats (120 to 140 g) were procured from Beijing Vital River Laboratory Animal Technology. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Shandong University (No. 2022-127). A rat model of T2DM was established as previously described [16]. Control rats were fed a normal chow diet (Supplementary Table 1) and received intraperitoneal injection of citrate buffer alone. The oral glucose tolerance test (OGTT) and homeostatic model assessment for insulin resistance (HOMA-IR) index were performed as previously reported protocols [17,18].
The interscapular BAT of rats was separated and digested with collagenase (Supplementary Table 2) as previously described [19]. The cell suspension was filtered sequentially through a 100- and 40-μm cell strainer (Solarbio, Beijing, China) to remove tissue and cell debris and then used for scRNA-seq. The brown adipocyte progenitor cells (BAPCs) from SVF of BAT (CP-R228, Procell Life, Wuhan, China) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% newborn calf serum (NCS, Biological Industries, BeitHaemek, Israel), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Grand Island, NY, USA). MACs were cultured in DMEM with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin for different studies.
Libraries for scRNA-seq were constructed using the 10× Genomics platform and sequenced on an Illumina HiSeq 2500 (San Diego, CA, USA) as previously described [20]. Cell Ranger and Seurat were used for quality control, data clustering, visualization, annotation and identification of marker genes. CellChat (http://www.cellchat.org) was used to analyze cell–cell communication/crosstalk between populations. Trajectory analysis was conducted using Monocle 2 (http://cole-trapnell-lab.github.io/monocle-release). Details of above data analysis procedures were presented in the Supplementary Methods.
Tissue samples were fixed, embedded in paraffin, sectioned, and deparaffinized. After blocking endogenous peroxidase activity, the sections were incubated with specific antibodies for immunofluorescence (IF) or immunohistochemistry (IHC). Detection was performed using either fluorescent secondary antibodies (Proteintech, Rosemont, IL, USA) for IF, or secondary antibodies and a 3,3´ diaminobenzidine (DAB) peroxidase substrate kit (Golden Bridge Biotechnology, Beijing, China) for IHC (Supplementary Table 3).
Cells were washed, fixed and incubated with an Oil Red O solution (Sigma, St. Louis, MO, USA). After washing, cells were visualized under a microscope. Densitometric quantification of the Oil Red O staining was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cells or tissues were washed and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China). Purified proteins were quantified, electrophoresed and transferred to polyvinylidene fluoride membranes (Millipore, Burlington, MA, USA). Membranes were blocked with 5% nonfat milk and sequentially incubated with primary and secondary antibodies (Supplementary Table 3). Finally, protein bands were detected using an enhanced chemiluminescence kit (Millipore). Densitometry analysis was performed using ImageJ software.
Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Quantitative polymerase chain reaction was conducted sequentially using PrimeScript RT Master Mix and TB Green Premix Ex Taq II (TaKaRa, Kusatsu, Japan). Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) gene was used as the internal standard. The relative mRNA expression levels were quantified using the 2-ΔΔCt method. All used primers were listed in Supplementary Table 4.
According to the manufacturer’s protocol, mitochondrial quantity and mitochondrial oxygen consumption underwent separate testing using Mitotracker Red (Beyotime) and an Extracellular Oxygen Consumption Assay Kit (Abcam, Cambridge, UK). For the latter, the slope values represented the oxygen consumption rate of each group.
Data were presented as mean±standard error of the mean/standard deviation or median and interquartile range. The Kolmogorov-Smirnov test was used to assess data normality. For normally distributed data, a two-tailed Student’s t-test or one-way analysis of variance (ANOVA) with an appropriate post hoc test was used for two-group or multiple-group comparisons, respectively. The Mann-Whitney U test was used for two-group comparisons of non-normally distributed data. Fisher’s exact test with Benjamini-Hochberg false discovery rate multiple-test correction was used for functional enrichment analysis. Statistical analysis was conducted using GraphPad Prism v8.1 (GraphPad Software Inc., San Diego, CA, USA) and R software (R Foundation for Statistical Computing, Vienna, Austria). A P value, or an adjusted P value, less than 0.05 was considered statistically significant. All experiments were independently repeated at least three times.
To reveal the cell types and functions of BAT as well as their changes in T2DM, we analyzed the SVF of BAT in the interscapular region of normal rats (NC) and T2DM rats by scRNA-seq. The results, including body weight, OGTT, fasting blood glucose level, serum insulin level, and HOMA-IR index, confirmed the successful establishment of the T2DM rat model (Supplementary Fig. 1). A total of 11,556 cells were sequenced, identifying four types of non-immune cells and five types of immune cells in the BAT SVF of both groups (Fig. 1A). Non-immune cells included ASPCs (Cd34, 5’-nucleotidase ecto [Nt5e]/Cd73, alanyl aminopeptidase, membrane [Anpep]/Cd13), fibroblasts (FBs) (fibronectin 1 [Fn1], discoidin domain receptor tyrosine kinase 2 [Ddr2]), smooth muscle cells (SMCs) (actin alpha 2 [Acta2], caldesmon 1 [Cald1]), and endothelial cells (ECs) (cadherin 5 [Cdh5], kinase insert domain receptor [Kdr]) (Fig. 1B). Notably, the expression profiles of the marker genes of ASPCs and SMCs were similar, suggesting that they may transdifferentiate into each other or have a similar origin (Supplementary Fig. 2). The immune cells included MACs (Cd68, C-type lectin domain containing 10A [Clec10a]), T cells (Cd3d, Cd3g), B cells (Cd19, Jchain), natural killer (NK) cells (granzyme M [Gzmm], perforin 1 [Prf1]), and dendritic cells (DCs) (fms related receptor tyrosine kinase 3 [Flt3], integrin subunit alpha X [Itgax]) (Fig. 1C). Moreover, the expression profiles of marker genes of MACs and DCs were also similar, indicating a possible close relationship between them (Supplementary Fig. 2). ASPCs and MACs were respectively the most abundant non-immune and immune cells in the normal and T2DM groups. While the cell types in BAT of T2DM rats did not change, the proportion of many cell types was altered compared to NC (Fig. 1D).
Functional enrichment analysis showed that the differentiation of vascular smooth muscle cells and adipocytes, angiogenesis and lipid metabolism were significantly enriched in ASPCs. The functions of FBs and SMCs were mainly involved in collagen metabolism, angiogenesis, extracellular matrix organization, the differentiation of chondrocytes, osteoblasts and glial cells, as well as SMC proliferation and migration. ECs primarily participated in angiogenesis, SMC proliferation, lipid metabolism, and mesenchymal cell differentiation (Fig. 1E). Regarding immune cells, the functions of T cells, B cells, NK cells, and DCs were all associated with immune and inflammation. In addition to immune functions, MACs were also involved in various non-immune functions, such as the regulation of non-immune cell differentiation, angiogenesis, and lipid metabolism (Fig. 1F).
Intercellular ligand-receptor analysis revealed significantly altered crosstalks between most cell types in the BAT SVF of T2DM rats versus controls, with 16 key signaling pathways notably changed. For example, inflammation-related pathways such as macrophage inhibitory factor (MIF) and interleukin 1 (IL-1) were markedly enhanced, and angiogenesis-related pathways like platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) were opened up (Fig. 2A-C, Supplementary Fig. 3A). Angiopoietin-like protein (ANGPTL) signaling was enhanced between ASPCs, ECs, and SMCs; MIF signaling was increased between almost all cell types (Fig. 2D and E). In T2DM rats, the main outgoing information flow changed from FBs to multiple cell types including ASPCs, SMCs, and FBs, and the main outgoing signaling changed from ANGPTL signaling to multiple pathways. The inflammatory IL-1 signaling outgoing was upregulated in ASPCs, and many angiogenic signals including colony stimulating factor, VEGF, and PDGF signaling were opened up in FBs (Fig. 2F). The cell types of main incoming information flow changed from DCs to multiple cell types, such as FBs, DCs, and SMCs, and the changes in signaling were similar to the outgoing signaling patterns (Fig. 2G).
In response to certain stimuli, cells in tissues can exhibit universally conserved responses independent of cell types. We found 44 genes with increased expression in all cell types of the BAT SVF in T2DM rats (Supplementary Fig. 3B). These genes were involved in various functions, including tissue remodeling, angiogenesis, reactive oxygen species (ROS) and inflammation, as well as important pathways such as IL-17, tumor necrosis factor (TNF) pathways (Supplementary Fig. 3C and D). ECs exhibited most transcription factor (TF) activity, including Sox17/18/7 for arteriogenesis/lymphangiogenesis (Supplementary Fig. 3E and F). Additionally, some highly active TFs, such as KLF transcription factor 7 (Klf7) which regulates insulin/adipogenesis, emerged in the BAT of T2DM rats (Supplementary Fig. 3F and G) [21].
ASPCs are predominantly pluripotent cells in adipose tissue. We found that ASPCs could be divided into the ASPC1 cluster and the T2DM-specific ASPC2 cluster (Fig. 3A). Pseudotime analysis showed that ASPC1 and ASPC2 were located at the same position in differentiation track branch 1, suggesting that they might have similar origins (Fig. 3B). ASPC1 could be subdivided into four subsets: Cyr61+ASPC1-s0 was mainly involved in inflammatory responses. Nt5e+ASPC1-s1 was associated with the differentiation of various cells, such as osteoblasts, myoblasts, and mesenchymal stem cells, reflecting the pluripotency of ASPC1-s1 and suggesting that ASPC1-s1 might be a classical ASPCs subset. However, in T2DM rats, the genes related to cell differentiation in the ASPC1-s1 subset were downregulated (Fig. 3C-F). The IF results also showed that in BAT-resident ASPCs from T2DM rats, the levels of markers (growth differentiation factor 10 [GDF10], odd-skipped related transcription factor 1 [OSR1], ERBB receptor feedback inhibitor 1 [ERRFI1], and collagen type I alpha 1 chain [COL1A1]) associated with differentiation potential to osteoblasts, myoblasts, epidermal cells and mesenchymal cells were significantly reduced (Supplementary Fig. 4) [22-26]. These results indicated the reduced pluripotency of ASPCs in BAT. Additionally, C-C motif chemokine ligand 4 (Ccl4)+ASPC1-s2 was associated with lymphocyte migration and chemotaxis. Collagen type III alpha 1 chain (Col3a1)+ASPC1-s3 was involved in extracellular matrix and collagen fibril organization (Fig. 3C-E). The pseudotime analysis also suggested that the classical ASPCs subcluster ASPC1-s1 gradually differentiates into the inflammatory ASPC1-s2 subcluster in T2DM rats (Fig. 3G).
The T2DM-specific ASPC2 cluster had five subsets: pentraxin 3 (Ptx3)+ASPC2-s0 was mainly associated with angiogenesis and SMC proliferation; Cd163+ASPC2-s1 was involved in neutrophil chemotaxis and myeloid differentiation; S100 calcium binding protein A9 (S100a9)+ASPC2-s2 was associated with chronic inflammation and neutrophil chemotaxis; Anpep+ ASPC2-s3 was involved in EC proliferation and adipocyte differentiation. Fc epsilon receptor Ig (Fcer1g)+ASPC2-s4 was related to TNF production and antigen presentation (Fig. 3H-J). Pseudotime analysis showed that during T2DM development, the T2DM-specific ASPC2 gradually differentiated from the ASPC2-s3 subset, which has proliferative and differentiation potential, to the ASPC2-s0 and ASPC2-s1 subsets with predominantly angiogenic and inflammatory functions (Fig. 3K).
Our results showed that ECs of BAT were divided into three subclusters (Fig. 4A). The apelin receptor (Aplnr)+Kdr+EC-s0 subcluster was closely related to angiogenesis, EC proliferation and differentiation, and VEGF response, indicating that EC-s0 might be classical ECs. Integral membrane protein 2A (Itm2a)+ leucine rich repeat containing 8 VRAC subunit C (Lrrc8c)+ECs1 was involved in brown adipocyte differentiation and SMC migration. IF further confirmed the presence of the EC-s1 subcluster in BAT (Fig. 4B-D). Fibulin 5 (Fbln5)+hypoxia inducible factor 1 subunit alpha (Hif1a)+apoprotein E (Apoe)+scavenger receptor class B member 1 (Scarb1)+EC-s2 was closely associated with the metabolism and transport of lipids, such as high-density lipoprotein, steroids, and triglycerides (Fig. 4B and C, Supplementary Fig. 5A).
The proportion of the three EC subclusters in the BAT of T2DM rats did not change significantly (Supplementary Fig. 5B). However, the expression of many genes was markedly altered. In the EC-s0 subcluster, the upregulated genes indicated enhanced granulocyte chemotaxis and regulation of vasculature development, while the downregulated genes suggested a reduction in the negative regulation of cell migration (Fig. 4E and F). In the EC-s1 subcluster, the upregulated genes suggested enhanced brown adipocyte differentiation and angiogenesis, whereas the downregulated genes denoted a decreased ability to respond to hypoxia and ROS (Fig. 4E and F). In the EC-s2 subcluster, the significant upregulation of Ccl2, C-X-C motif chemokine ligand 1 (Cxcl1), Ccl5, Ccl4, and insulin like growth factor 1 (Igf1) expression suggested increased neutrophil chemotaxis and SMC proliferation, while the downregulation of fatty acid binding protein 5 (Fabp5), caveolin 1 (Cav1), peroxisome proliferator-activated receptor γ (Pparg), superoxide dismutase 1 (Sod1), and Scarb1 expression indicated that the functions of regulation of lipid metabolism, cholesterol storage, and low-density lipoprotein particle clearance were diminished (Fig. 4E and F).
We found that the MACs of BAT had six subclusters (Fig. 5A). The EGF like domain multiple 7 (Egfl7)+MAC-s0 subcluster was closely associated with the immune responses, angiogenesis, SMC proliferation and EC function. Rarres2+MAC-s1 was involved in major histocompatibility complex (MHC) class II antigen presentation, proliferation/differentiation of multiple immune cells (T cells, B cells, DCs, and MACs) and brown adipocyte differentiation. RT1 class II, locus Bb (RT1-Bb)+MACs2 primarily participated in antigen presentation via MHC II and γ-interferon responses. Fc gamma receptor IIIa (Fcgr3a)+MAC-s3 was related to MHC class I antigen presentation and immune effector processes. Interleukin 1 receptor type 2 (Il1r2) +MAC-s4 was implicated in granulocyte chemotaxis and ROS metabolism. Ficolin B (Fcnb)+MAC-s5 functioned in phagocytosis and cytokine secretion (Fig. 5B and C). Moreover, the classical marker genes of M1 and M2 MACs were expressed in multiple MAC subclusters identified from the scRNA-seq data (Supplementary Fig. 5C and D). These results indicated that MAC heterogeneity was more complex than previously thought.
In the BAT of T2DM rats, the percentage of the MAC-s0 subcluster increased, while the percentages of the MAC-s1, MAC-s3, and MAC-s4 subclusters decreased (Fig. 5D). The expression levels of many genes were changed in each subcluster. In the MAC-s0 subcluster, the upregulated genes suggested enhanced acute inflammatory responses and angiogenesis, while the downregulated genes indicated a diminished Fc gamma receptor pathway. In the MAC-s1 subcluster, the upregulated genes illustrated an enhanced positive regulation of adipocyte differentiation and monocyte chemotaxis, whereas the downregulated genes implied a decrease in differentiation of T cells, lymphocytes and DCs, and smooth muscle relaxation (Fig. 5E and F). In the MAC-s2 subcluster, the upregulation of Ccl5, Cxcl9, and Cxcl3 expression implied an enhanced response to transforming growth factor-β (TGF-β) stimulation and chemokine-mediated signaling, while the downregulated genes demonstrated a decrease in leukocyte activation regulation. In the MAC-s3 subcluster, the significantly changed genes suggested an enhanced response to IL-1 and a reduction in ROS metabolic regulation. In the MAC-s4 subcluster, the significantly altered genes denoted an enhanced response to TGF-β and PDGF and a decrease in cell killing ability. Finally, in the MAC-s5 subcluster, the significantly regulated genes indicated enhanced regulation of SMC proliferation and a decrease in ROS metabolic regulation (Fig. 5E and F).
Our results suggested that Rarres2+MACs, which were reduced in the BAT of T2DM rats, might promote brown adipocyte differentiation. IF results confirmed the presence of Rarres2+ MACs in BAT (Fig. 6A). Moreover, the percentage of Rarres2+ MACs in WAT MACs was significantly lower than that in BAT MACs (Fig. 6B, Supplementary Fig. 6A). Rarres2 expression was also higher in BAT-derived SVF than in WAT-derived SVF and in brown adipocytes (Fig. 6C). Additionally, Rarres2 was barely expressed in peritoneal MACs, but mainly expressed in MACs of BAT (Supplementary Fig. 6B and C). These results suggested tissue specificity of Rarres2+MACs in BAT-derived SVF. BAPCs cocultured with Rarres2+MACs had a more mature morphology and more lipid droplets in their cytoplasm (Fig. 6D and E). Compared to BAPCs without treatment or with Rarres2-MACs treatment, BAPCs treated with Rarres2+ MACs expressed higher levels of PR domain-containing 16 (PRDM16) and uncoupling protein 1 (UCP1) proteins, indicating enhanced BAPC differentiation. Rarres2+MACs also significantly upregulated the expression of cytochrome c oxidase subunit 7A2 like (COX7RP) and transcription factor A, mitochondrial (TFAM) proteins, two key factors in mitochondrial functions (Fig. 6F) and increased the number and oxygen consumption of mitochondria (Fig. 6G and H, Supplementary Fig. 6D and E), suggesting that Rarres2+MACs effectively promoted energy metabolism in BAPCs. Meanwhile, anti-inflammatory Rarres2-CD206+M2 MACs did not significantly affect the differentiation and function of BAPCs, which indicated that the unique roles of Rarres2+MAC mentioned above were not a general function of anti-inflammatory MACs (Supplementary Fig. 6F and G). Rarres2 has been reported to bind the membrane receptor chemerin chemokine-like receptor 1 (Cmklr1). We found that CMKLR1 was highly expressed in both BAPCs and BAT (Supplementary Fig. 7A). Moreover, Rarres2+MACs significantly increased the levels of phosphorylated mammalian target of rapamycin (p-mTOR)/mTOR, p-AKT/AKT, and phosphorylated extracellular signal-regulated kinase (p-ERK)/ERK, indicating enhanced phosphorylation levels of mTOR, AKT, and ERK, which are downstream molecules of the Rarres2-Cmklr1 axis. These results implied the activation of AKT-mTOR and ERK pathway. Furthermore, Rarres2+MACs significantly enhanced the expression of TFs CCAAT enhancer binding protein beta (CEBPβ), CEBPα, and PPARγ, which play crucial roles in the differentiation of adipose precursor cells (Fig. 6I). However, the increased expression of CEBPβ, CEBPα, and PPARγ was effectively eliminated by AKT-siRNA, mTOR-siRNA, and ERK-siRNA. Moreover, the upregulated expression of CEBPα and PPARγ was markedly suppressed by CEBPβsiRNA (Supplementary Fig. 7B-E). These findings suggested that Rarres2+MACs regulated brown adipocytes through adipocyte-immune crosstalk consisting of the Rarres2-Cmklr1 axis (Supplementary Fig. 8).
MACs play key roles in adipose tissue inflammation and diabetes [27]. M1 MACs are thought to enhance inflammation and insulin resistance, thus exacerbating T2DM development, while M2 MACs have a protective anti-inflammatory role. However, recent scRNA-seq studies have shown greater heterogeneity in MACs than previously known. Traditional M1 and M2 markers coexist in many MAC subclusters identified by scRNA-seq [28]. Moreover, MACs have numerous non-immune functions [29]. We found that Egfl7+MACs (MAC-s0) and Rarres2+MACs regulated angiogenesis, and immune and non-immune cell proliferation and differentiation. The expression of these function-related genes was significantly upregulated in T2DM rats. Additionally, the downregulation of Fc-γ receptor pathway and genes positively regulating monocyte chemotaxis indicated changes in the immune and phagocytic function of MAC-s0 [30]. These findings underscore that the types and functions of MACs in BAT are intricate. Further in-depth studies are needed to decipher the functional regulation of BAT, as well as the complex interaction between immunity and metabolism.
To date, research on MACs of BAT remains limited. Gallerand et al. [31] identified four MAC subclusters in mouse BAT: one subcluster with classical immune functions, two subclusters regulating lipid metabolism and BAT expansion, and another subcluster involved in tissue remodeling. Our study identified a novel Rarres2+MAC subcluster that regulated the differentiation of multiple cell types, including brown adipocytes. This subcluster exhibits an important metabolic regulatory function acting as a bridge between metabolism and immunity. Our analysis of a published dataset [32] also showed that Rarres2+MAC population in BAT was increased in cold conditions. Rarres2 has been demonstrated to bind the membrane receptor Cmklr1 and be expressed in both WAT and BAT, playing critical roles in white and brown adipocyte differentiation and energy metabolism [33]. Our findings showed that they were abundant in BAT but rare in WAT, indicating significant tissue specificity. The main downstream pathways of the Rarres2/Cmklr1 axis involved in promoting the differentiation of 3T3-L1 cells and human adipocytes include Akt-mTOR and ERK pathways [34]. Both pathways can regulate the downstream adipogenic TFs CEBPβ, PPARγ, and CEBPα [35-38]. Zhang et al. [33] reported reduced brown adipocyte differentiation and mitochondrial content and function in Rarres2-/- mice. Similarly, we found that Rarres2+MACs promoted BAPCs differentiation and increased mitochondrial number and function.
ASPCs are key pluripotent cells in adipose tissues. Using scRNA-seq, Vijay et al. [39] identified five subclusters in ASPCs of WAT, two of which expressed adipose stem cell markers, similar to ASPC1-s1 in our study. Another subcluster expressed genes related to fibrosis and extracellular matrix, similar to ASPC1-s3 in our study. Subcluster 5 expressed high levels of inflammatory markers, functioning similarly to ASPC1-s0 and ASPC1-s2 in our study. Some studies reported that WAT-derived ASPCs had various metastable states that could be perturbed toward adipogenic or non-adipogenic fates [40,41]. We found that in diabetic rats, the multidirectional differentiation potential of ASPCs in BAT was markedly reduced, likely due to increased inflammation in BAT.
ScRNA-seq studies have revealed that aortic ECs are divided into different subsets [42]. We found that ECs in BAT also form several subclusters, including lipid metabolism-related ECs. These ECs specifically expressed lipoprotein lipase (Lpl), Apoe, and Scarb1, suggesting a role in lipid transport and storage, which was consistent with a recent report [43]. We identified a novel Lrrc8c+EC subset possibly involved in brown adipocyte differentiation and resemble ECs previously reported to promote adipose precursor cell differentiation in WAT [44]. These studies, including ours, highlight the heterogeneity of ECs and their important roles in adipocyte differentiation. Moreover, we found that ECs also produced chemokines and inflammatory factors, contributing to BAT inflammation.
BAT is known to burn fat through thermogenesis and its dysfunction significantly contributes to the development of many metabolic diseases [45]. Our results revealed the presence of many inflammatory cells in BAT. Moreover, almost all cells in the BAT SVF of diabetic rats highly expressed genes that promote inflammation and angiogenesis. These findings suggest that although BAT in healthy individuals has beneficial effects on glucose, lipid and energy metabolism, which are diminished in pathological states such as diabetes. Increased angiogenesis and inflammation make BAT a significant source of proinflammatory factors and cells, which exacerbates systemic inflammation and metabolic abnormalities, facilitating T2DM development.
In conclusion, we revealed thze cellular composition and functional characteristics of the BAT SVF and their abnormalities in T2DM at a single-cell resolution. We also identified a novel Rarres2+MAC subset that promoted brown adipocyte differentiation and function. However, our study still has some shortcomings. For instance, the function and clinical significance of Rarres2+MACs require further investigation using animal models and clinical samples. Nevertheless, our findings shed light on the role of BAT in T2DM pathogenesis and offer insights into its mechanism, which could have implications for future therapeutic strategies.
SUPPLEMENTARY MATERIALS
Supplementary materials related to this article can be found online at https://doi.org/10.4093/dmj.2023.0278.
Supplementary Table 2.
The composition of the collagenase buffer
Supplementary Table 3.
The antibodies used in the present study
Supplementary Table 4.
The primer sequences used for real-time polymerase chain reaction
Supplementary Fig. 1.
Assessment of type 2 diabetes mellitus (T2DM) rat model. (A) Comparison of body weight of normal rats (NC) and T2DM rats during the experiment period. (B) Comparison of oral glucose tolerance test results in NC and T2DM rats. (C-E) Comparison of the levels of fasting blood glucose (C), serum insulin (D), and homeostatic model assessment for insulin resistance (HOMA-IR) index (E) in NC and T2DM rats. A two-tailed Student’s t-test was used for statistical analysis. aP<0.01, bP<0.001, cP<0.0001.
Supplementary Fig. 2.
Heatmap of relative expression levels of the top 20 marker genes for each cell subpopulation in the brown adipose tissue stromal vascular fraction. ASPC, adipose stem/progenitor cell; FB, fibroblast; SMC, smooth muscle cell; EC, endothelial cell; MAC, macrophage; NK, natural killer; DC, dendritic cell.
Supplementary Fig. 3.
Crosstalk strength between different cell types, conserved responses and transcription factor (TF) activity in brown adipose tissue (BAT) stromal vascular fraction (SVF) of type 2 diabetes mellitus (T2DM) rats. (A) Heatmap of the crosstalk strength between various cell types in the BAT SVF of normal rats (NC) (left panel) and T2DM rats (right panel). The top (right) bar indicates the sum of crosstalk strength in corresponding column (row) in the heatmap. Crosstalk strength: the number/probability of crosstalks among different cell populations calculated by Cellchat. (B-D) Heatmap of fold changes in the expression levels of 44 significantly upregulated genes in all nine cell types of BAT in T2DM rats compared to the NC (B) and their enrichment analysis in the Gene Ontology (GO) Term (C) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (D). (E, F) Heatmap of viper scores of the top 20 highly active TFs in each cell type of BAT SVF in NC rats (E) and T2DM rats (F). Viper scores: evaluation of TF activity according to virtual inference of protein-activity by enriched regulon analysis. (G) Functional enrichment analysis of the target genes regulated by the TF Klf7 in endothelial cells (ECs). ASPC, adipose stem/progenitor cell; FB, fibroblast; SMC, smooth muscle cell; MAC, macrophage; NK, natural killer; DC, dendritic cell; IL, interleukin; TNF, tumor necrosis factor; AGE-RAGE, advanced glycation end products-receptor for advanced glycation end products; NF, nuclear factor; NOD, ncleotide oligomerization domain.
Supplementary Fig. 4.
Expression levels of markers associated with the differentiation potential in brown adipose tissue-resident adipose stem/progenitor cells. (A-D) Representative immunofluorescence images of (A) growth differentiation factor 10 (GDF10), (B) odd-skipped related transcription factor 1 (OSR1), (C) ERBB receptor feedback inhibitor 1 (ERRFI1), and (D) collagen type I alpha 1 chain (COL1A1) protein expression in normal rats (NC) and type 2 diabetes mellitus (T2DM) rats (left panel) and their quantitative comparison (right panel). A two-tailed Student’s t-test was used for statistical analysis. DAPI, 4´,6-diamidino-2-phenylindole. aP<0.01, bP<0.001.
Supplementary Fig. 5.
Marker genes and proportion of three endothelial cell (EC) subsets in ECs population and the marker gene expression in different macrophage (MAC) subsets. (A) Violin plots of the relative expression levels of apoprotein E (Apoe) and scavenger receptor class B member 1 (Scarb1) in various subsets of ECs. (B) Proportion of each EC subset in the total ECs of brown adipose tissue in normal rats (NC) and type 2 diabetes mellitus (T2DM) rats. (C, D) Violin plots of relative expression levels of classical marker genes of M1 (C) and M2 (D) MACs in each MAC subcluster identified by single-cell RNA sequencing. Il1b, interleukin 1 beta; Fcgr1a, Fc gamma receptor Ia; Fcgr2b, Fc gamma receptor IIb; Egfl7, EGF like domain multiple 7; Rarres2, retinoic acid receptor responder 2; RT1-Bb, RT1 class II, locus Bb; Fcgr3a, Fc gamma receptor IIIa; Il1r2, interleukin 1 receptor type 2; Fcnb, ficolin B; Mrc1, mannose receptor, C type 1; Folr2, folate receptor beta; Tgfb1, transforming growth factor-β1.
Supplementary Fig. 6.
Distribution of retinoic acid receptor responder 2 (Rarres2)+macrophage (MAC) in different adipose tissues and the effect of Rarres2+MAC or M2 MACs on the differentiation and function of brown adipocytes. (A) Representative immunofluorescence images of CD68+Rarres2+MACs in white adipose tissue (WAT) and brown adipose tissue (BAT). (B) Comparison of relative expression levels of Rarres2 mRNA in peritoneal MACs and BAT MACs. (C) Violin plots of relative expression levels of Rarres2 mRNA in each cell clusters of the BAT stromal vascular fraction. (D) Representative immunofluorescence images of mitochondria stained by Mitotracker Red in brown adipocyte progenitor cells (BAPCs) with different treatments. (E) Relative oxygen consumption of BAPCs with different treatments at multiple time points compared to those of corresponding control group (i.e., signal control wells added phosphate buffered saline and O2 consumption reagent). (F) Representative Western blot images of differentiation-related proteins and mitochondrial functional proteins in BAPCs with or without M2 MAC treatment (left panel) and their quantitative comparison (right panel). Fold change=The relative expression levels of each group compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH)/the mean of relative expression levels of D0 BAPC compared to GAPDH. (G) Relative oxygen consumption at multiple time points compared to the corresponding signal of control group (namely signal control wells added PBS and O2 consumption reagent) at the same time point (left panel) and the comparison of the oxygen consumption rate (OCR) at the plateau phase (right panel) of BAPCs with or without M2 MAC treatment. Fold change=The OCRs of each group/the mean of OCRs of D0 BAPC. Two-tailed Student’s t-test (B) and one-way analysis of variance (ANOVA) test with appropriate correction (E-G) were used for statistical analysis. ASPC, adipose stem/progenitor cell; FB, fibroblast; SMC, smooth muscle cell; EC, endothelial cell; NK, natural killer; DC, dendritic cell; DAPI, 4´,6-diamidino-2-phenylindole; UCP1, uncoupling protein 1; COX7RP, cytochrome c oxidase subunit 7A2 like; CEBPα, CCAAT enhancer binding protein α; TFAM, transcription factor A, mitochondrial; PRDM16, PR domain-containing 16; NS, no significance. aP<0.01, bP<0.001, cP<0.0001.
Supplementary Fig. 7.
Expression levels of chemerin chemokine-like receptor 1 (CMKLR1) in brown adipocyte progenitor cells (BAPCs) and brown adipose tissue (BAT) and key mechanistic molecules in BAPCs with different treatments. (A) Representative immunofluorescence images of CMKLR1 protein expression in BAPCs (upper panel) and BAT (down panel). (B-E) Representative Western blot images of key signaling molecules involved in the differentiation and function in BAPCs treated with (B) AKT-siRNA, (C) mammalian target of rapamycin (mTOR)-siRNA, (D) extracellular signal-regulated kinase (ERK)-siRNA, or (E) CCAAT enhancer binding protein beta (CEBPβ)-siRNA (left panel) and their quantitative comparison (right panel). Fold change=The relative expression levels of each group compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH)/the mean of relative expression levels of D8 BAPC compared to GAPDH. One-way analysis of variance (ANOVA) test with appropriate correction (B-E) was used for statistical analysis. DAPI, 4´,6-diamidino-2-phenylindole; PPARγ, peroxisome proliferator-activated receptor γ; MAC, macrophage; Si-NC, negative control-siRNA. aP<0.05, bP<0.01, cP<0.001, dP<0.0001.
Supplementary Fig. 8.
Schematic representation of the mechanism by which retinoic acid receptor responder 2 (Rarres2)+macrophages (MACs) promote differentiation and function of brown adipocytes. CMKLR1, chemerin chemokine-like receptor 1; mTOR, mammalian target of rapamycin; ERK, extracellular signal-regulated kinase; CEBPβ, CCAAT enhancer binding protein beta; PPARγ, peroxisome proliferator-activated receptor γ.
Notes
REFERENCES
1. Brandon AE, Liao BM, Diakanastasis B, Parker BL, Raddatz K, McManus SA, et al. Protein kinase C epsilon deletion in adipose tissue, but not in liver, improves glucose tolerance. Cell Metab. 2019; 29:183–91.

2. Iwen KA, Backhaus J, Cassens M, Waltl M, Hedesan OC, Merkel M, et al. Cold-induced brown adipose tissue activity alters plasma fatty acids and improves glucose metabolism in men. J Clin Endocrinol Metab. 2017; 102:4226–34.

3. Villarroya F, Cereijo R, Villarroya J, Giralt M. Brown adipose tissue as a secretory organ. Nat Rev Endocrinol. 2017; 13:26–35.

4. Shimizu I, Walsh K. The whitening of brown fat and its implications for weight management in obesity. Curr Obes Rep. 2015; 4:224–9.

5. White JD, Dewal RS, Stanford KI. The beneficial effects of brown adipose tissue transplantation. Mol Aspects Med. 2019; 68:74–81.

6. Grunewald ZI, Winn NC, Gastecki ML, Woodford ML, Ball JR, Hansen SA, et al. Removal of interscapular brown adipose tissue increases aortic stiffness despite normal systemic glucose metabolism in mice. Am J Physiol Regul Integr Comp Physiol. 2018; 314:R584–97.

7. Cox N, Geissmann F. Macrophage ontogeny in the control of adipose tissue biology. Curr Opin Immunol. 2020; 62:1–8.

8. Rosina M, Ceci V, Turchi R, Chuan L, Borcherding N, Sciarretta F, et al. Ejection of damaged mitochondria and their removal by macrophages ensure efficient thermogenesis in brown adipose tissue. Cell Metab. 2022; 34:533–48.

9. Fischer K, Ruiz HH, Jhun K, Finan B, Oberlin DJ, van der Heide V, et al. Alternatively activated macrophages do not synthesize catecholamines or contribute to adipose tissue adaptive thermogenesis. Nat Med. 2017; 23:623–30.

10. Yang D, Yang L, Cai J, Hu X, Li H, Zhang X, et al. A sweet spot for macrophages: focusing on polarization. Pharmacol Res. 2021; 167:105576.

11. Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018; 155:407–17.

12. Mosser DM, Hamidzadeh K, Goncalves R. Macrophages and the maintenance of homeostasis. Cell Mol Immunol. 2021; 18:579–87.

13. Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018; 559:103–8.

14. Song A, Dai W, Jang MJ, Medrano L, Li Z, Zhao H, et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J Clin Invest. 2020; 130:247–57.

15. Nguyen HP, Lin F, Yi D, Xie Y, Dinh J, Xue P, et al. Aging-dependent regulatory cells emerge in subcutaneous fat to inhibit adipogenesis. Dev Cell. 2021; 56:1437–51.

16. Shao Q, Meng L, Lee S, Tse G, Gong M, Zhang Z, et al. Empagliflozin, a sodium glucose co-transporter-2 inhibitor, alleviates atrial remodeling and improves mitochondrial function in high-fat diet/streptozotocin-induced diabetic rats. Cardiovasc Diabetol. 2019; 18:165.

17. Tahan V, Yavuz D, Imeryuz N, Avsar E, Tozun N. Oral glucose tolerance deteriorates in rats fed with methionine choline deficient diet. J Hepatol. 2004; 41:352.

18. Todero J, Douillet C, Shumway AJ, Koller BH, Kanke M, Phuong DJ, et al. Molecular and metabolic analysis of arsenic-exposed humanized AS3MT mice. Environ Health Perspect. 2023; 131:127021.

19. Hu B, Jin C, Zeng X, Resch JM, Jedrychowski MP, Yang Z, et al. γδ T cells and adipocyte IL-17RC control fat innervation and thermogenesis. Nature. 2020; 578:610–4.

20. Gaydosik AM, Tabib T, Domsic R, Khanna D, Lafyatis R, Fuschiotti P. Single-cell transcriptome analysis identifies skin-specific T-cell responses in systemic sclerosis. Ann Rheum Dis. 2021; 80:1453–60.

21. Yang X, Liang M, Tang Y, Ma D, Li M, Yuan C, et al. KLF7 promotes adipocyte inflammation and glucose metabolism disorder by activating the PKCζ/NF-κB pathway. FASEB J. 2023; 37:e23033.

22. Brandt KJ, Burger F, Baptista D, Roth A, Fernandes da Silva R, Montecucco F, et al. Single-cell analysis uncovers osteoblast factor growth differentiation factor 10 as mediator of vascular smooth muscle cell phenotypic modulation associated with plaque rupture in human carotid artery disease. Int J Mol Sci. 2022; 23:1796.

23. Vallecillo-Garcia P, Orgeur M, Vom Hofe-Schneider S, Stumm J, Kappert V, Ibrahim DM, et al. Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nat Commun. 2017; 8:1218.

24. Kotsaris G, Qazi TH, Bucher CH, Zahid H, Pohle-Kronawitter S, Ugorets V, et al. Odd skipped-related 1 controls the pro-regenerative response of fibro-adipogenic progenitors. NPJ Regen Med. 2023; 8:19.

25. Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006; 12:568–73.

26. Comba A, Faisal SM, Dunn PJ, Argento AE, Hollon TC, AlHolou WN, et al. Spatiotemporal analysis of glioma heterogeneity reveals COL1A1 as an actionable target to disrupt tumor progression. Nat Commun. 2022; 13:3606.

27. Chavakis T, Alexaki VI, Ferrante AW Jr. Macrophage function in adipose tissue homeostasis and metabolic inflammation. Nat Immunol. 2023; 24:757–66.

28. Yu L, Zhang Y, Liu C, Wu X, Wang S, Sui W, et al. Heterogeneity of macrophages in atherosclerosis revealed by single-cell RNA sequencing. FASEB J. 2023; 37:e22810.

29. Burl RB, Ramseyer VD, Rondini EA, Pique-Regi R, Lee YH, Granneman JG. Deconstructing adipogenesis induced by β3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 2018; 28:300–9.

30. Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008; 8:34–47.
31. Gallerand A, Stunault MI, Merlin J, Luehmann HP, Sultan DH, Firulyova MM, et al. Brown adipose tissue monocytes support tissue expansion. Nat Commun. 2021; 12:5255.

32. Burl RB, Rondini EA, Wei H, Pique-Regi R, Granneman JG. Deconstructing cold-induced brown adipocyte neogenesis in mice. Elife. 2022; 11:e80167.

33. Zhang Y, Shen WJ, Qiu S, Yang P, Dempsey G, Zhao L, et al. Chemerin regulates formation and function of brown adipose tissue: ablation results in increased insulin resistance with high fat challenge and aging. FASEB J. 2021; 35:e21687.

34. Jiang Y, Liu P, Jiao W, Meng J, Feng J. Gax suppresses chemerin/CMKLR1-induced preadipocyte biofunctions through the inhibition of Akt/mTOR and ERK signaling pathways. J Cell Physiol. 2018; 233:572–86.

35. Roh SG, Song SH, Choi KC, Katoh K, Wittamer V, Parmentier M, et al. Chemerin: a new adipokine that modulates adipogenesis via its own receptor. Biochem Biophys Res Commun. 2007; 362:1013–8.
36. Lee M, Sorn SR, Lee Y, Kang I. Salt Induces Adipogenesis/lipogenesis and inflammatory adipocytokines secretion in adipocytes. Int J Mol Sci. 2019; 20:160.

37. Lowell BB. PPARgamma: an essential regulator of adipogenesis and modulator of fat cell function. Cell. 1999; 99:239–42.

38. Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, et al. The IRE1-alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009; 9:556–64.
39. Vijay J, Gauthier MF, Biswell RL, Louiselle DA, Johnston JJ, Cheung WA, et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat Metab. 2020; 2:97–109.

40. Yamamoto M, Nagasawa Y, Fujimori K. Glycyrrhizic acid suppresses early stage of adipogenesis through repression of MEK/ERK-mediated C/EBPβ and C/EBPδ expression in 3T3-L1 cells. Chem Biol Interact. 2021; 346:109595.

41. Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, et al. A PDGFRα-mediated switch toward CD9high adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metab. 2017; 25:673–85.

42. Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, et al. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation. 2019; 140:147–63.

43. Yu L, Dai Y, Mineo C. Novel functions of endothelial scavenger receptor class B type I. Curr Atheroscler Rep. 2021; 23:6.

Fig. 1.
Cell composition and function of brown adipose tissue (BAT) in normal rats (NC) and type 2 diabetes mellitus (T2DM) rats. (A) The t-distributed stochastic neighbor embedding (t-SNE) plot (left panel) and canonical correlation analysis (CCA) plot (right panel) of single-cell RNA sequencing data of the BAT stromal vascular fraction (SVF) in the NC and T2DM rats. (B, C) Violin plots of the relative expression levels of marker genes for non-immune (B) and immune (C) cell clusters in the BAT SVF. (D) Percentage of each cell cluster in non-immune cells (upper panel) or immune cells (lower panel) in BAT SVF of the NC rats or T2DM rats. (E, F) Functional enrichment analysis of non-immune cells (E) and immune cells (F) in BAT SVF. Fisher’s exact test with Benjamini-Hochberg false discovery rate multiple-test correction was used for the statistical analysis in E and F. ASPC, adipose stem/progenitor cell; FB, fibroblast; SMC, smooth muscle cell; EC, endothelial cell; MAC, macrophage; NK, natural killer; DC, dendritic cell; VSMC, vascular smooth muscle cell.
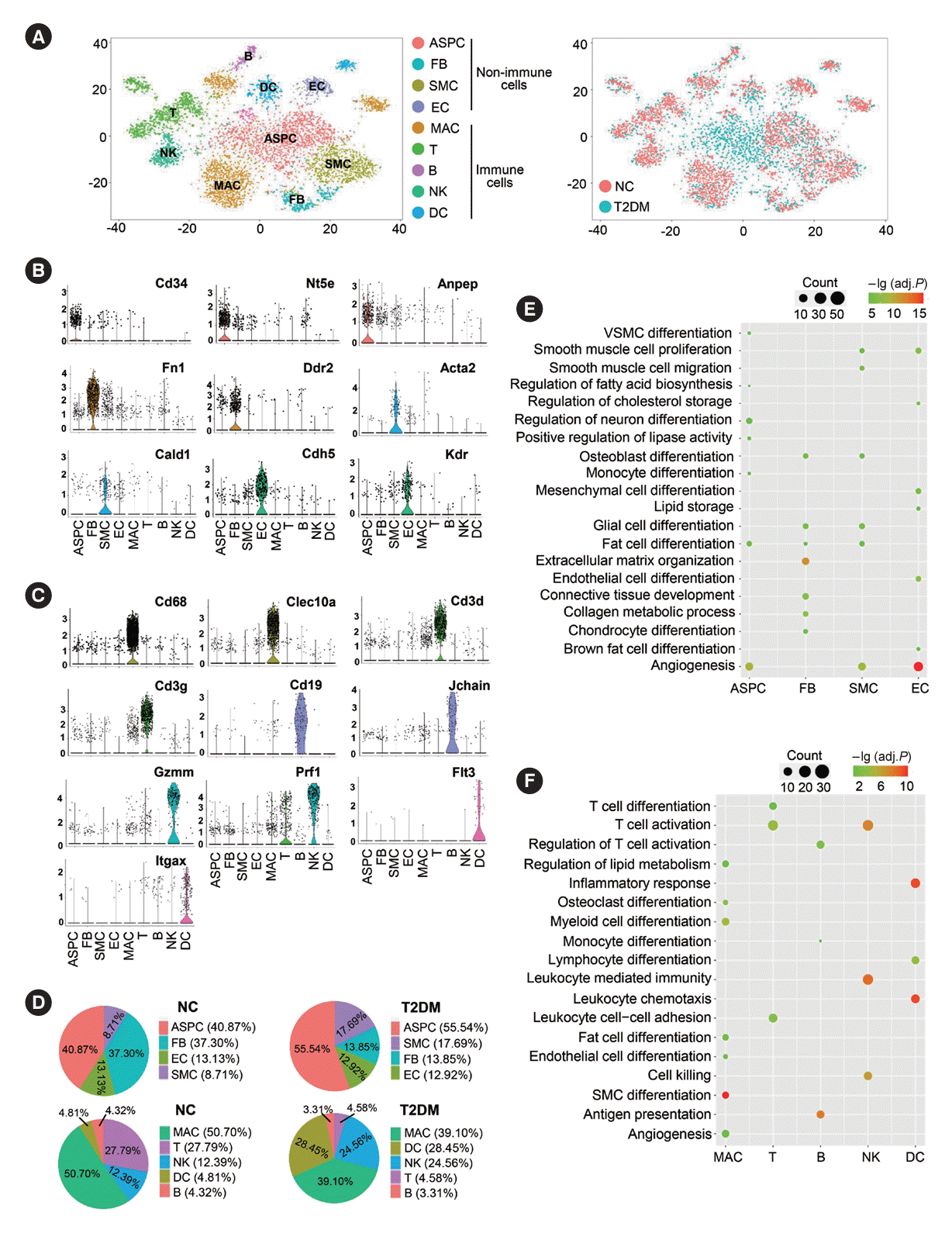
Fig. 2.
Enhanced intercellular crosstalk in brown adipose tissue (BAT) stromal vascular fraction (SVF) of type 2 diabetes mellitus (T2DM) rats. (A) Crosstalk numbers between each cell type in BAT SVF of normal rats (NC) and T2DM rats. Cells emitting arrows (indicated by arrows) express ligands (receptors). Edge width: crosstalk numbers. (B) Heatmap of the differential number of cell–cell interactions/crosstalks between distinct cell populations in BAT SVF of T2DM rats compared to NC rats. The top (right) bar shows the sum number of the differential interactions/crosstalks of T2DM rats compared to NC rats in corresponding column (row) in the heatmap. (C) Bar graph of the relative (left) and overall (right) information flow, which is defined by total communication probability among all pairs of cell populations in BAT SVF. (D, E) Chord diagram of the angiopoietin-like protein (ANGPTL) (D) and macrophage inhibitory factor (MIF) (E) signaling network between various cell types in BAT SVF. (F, G) Heatmap of the interaction/crosstalks strength of outgoing (F) and incoming (G) signaling pathways among various cell types. The top (right) bar presents the sum of the relative interaction/crosstalks strength of all (a) signaling pathway(s) in a (all) cell population(s). Relative strength: the contribution level of certain signal to outgoing or incoming signaling in a (all) cell population(s) calculated by Cellchat. ASPC, adipose stem/progenitor cell; FB, fibroblast; SMC, smooth muscle cell; EC, endothelial cell; MAC, macrophage; NK, natural killer; DC, dendritic cell; NT, neurotransmitter; CCL, chemokine C-C motif ligand; GAS, growth arrest-specific; APRIL, a proliferation-inducing ligand; IL, interleukin; MK, mitogen-activated protein kinase; PROS, protein S; PDGF, platelet-derived growth factor; CXCL, C-X-C motif chemokine ligand; BAFF, b-cell activating factor; VEGF, vascular endothelial growth factor; CSF, colony stimulating factor.
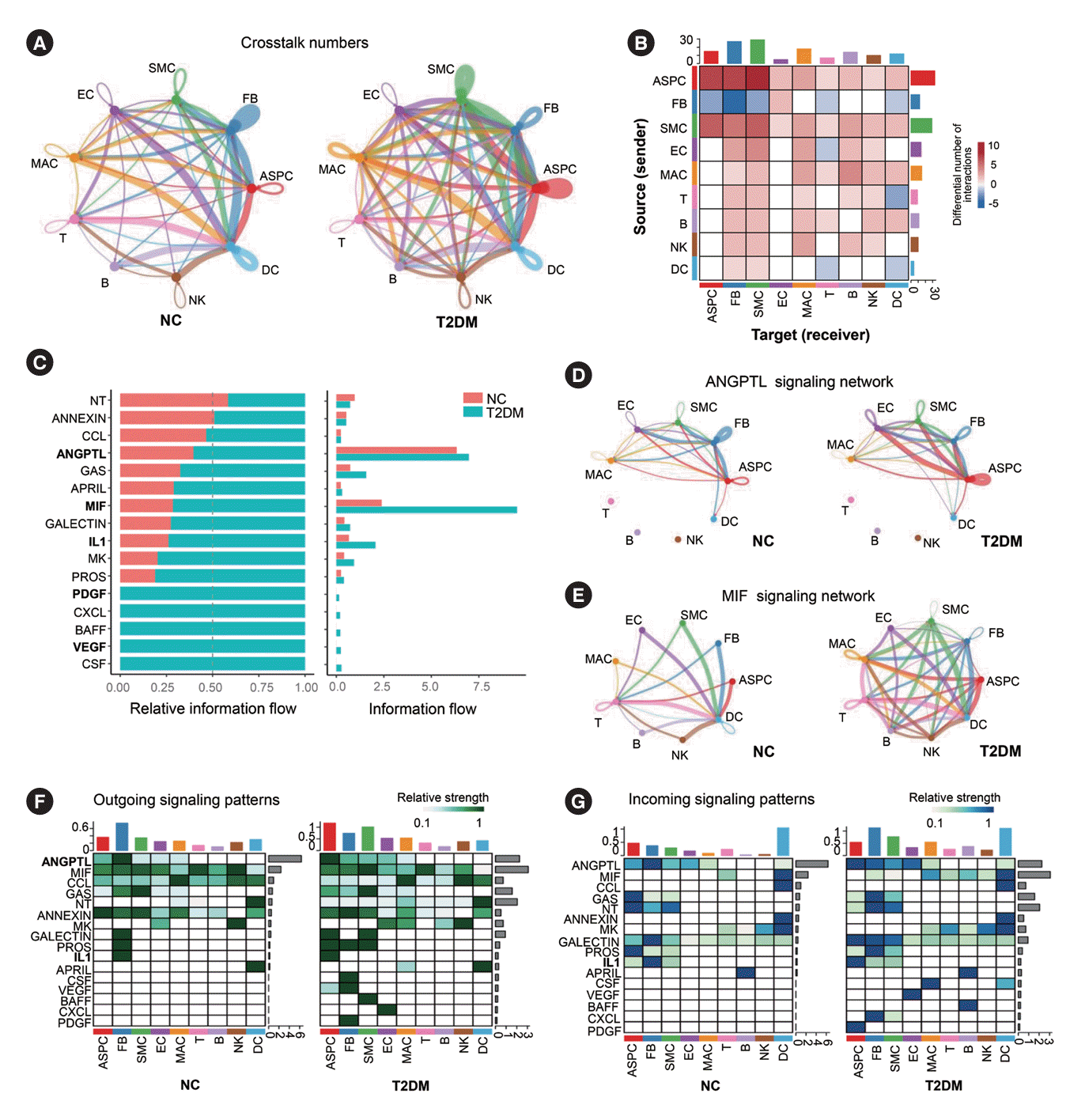
Fig. 3.
Subclusters and functions of adipose stem/progenitor cells (ASPCs) in brown adipose tissue (BAT). (A) The canonical correlation analysis (CCA) plot of ASPC subsets in BAT. (B) Distribution of ASPC1 and ASPC2 subsets on the pseudotime trajectory. (C, H) The t-distributed stochastic neighbor embedding (t-SNE) plot of the subclusters of ASPC1 (C) or ASPC2 (H) in BAT. (D, I) Violin plots of the relative expression levels of marker genes in each subpopulation of ASPC1 (D) or ASPC2 subcluster (I). (E, J) Functional enrichment analysis of the four subclusters of ASPC1 (E) or the five subclusters of ASPC2 (J). Circle size represents the number of related genes of each item. (F) Heatmap of relative expression levels and functional enrichment of representative differentially expressed genes in the ASPC1-s1 subcluster of ASPC1 in BAT of type 2 diabetes mellitus (T2DM) rats compared to normal rats (NC). (G) Distribution of ASPC1-s1 and ASPC1-s2 subclusters of ASPC1 on the pseudotime trajectory. (K) Distribution of ASPC2-s0, ASPC2-s1 and ASPC2-s3 subclusters of ASPC2 on the pseudotime trajectory. Fisher’s exact test with Benjamini-Hochberg false discovery rate multiple-test correction was used for the statistical analysis in E and J. The Mann-Whitney U test was performed for the statistical analysis in F. TNF, tumor necrosis factor; TGF-β, PDGF, platelet-derived growth factor.
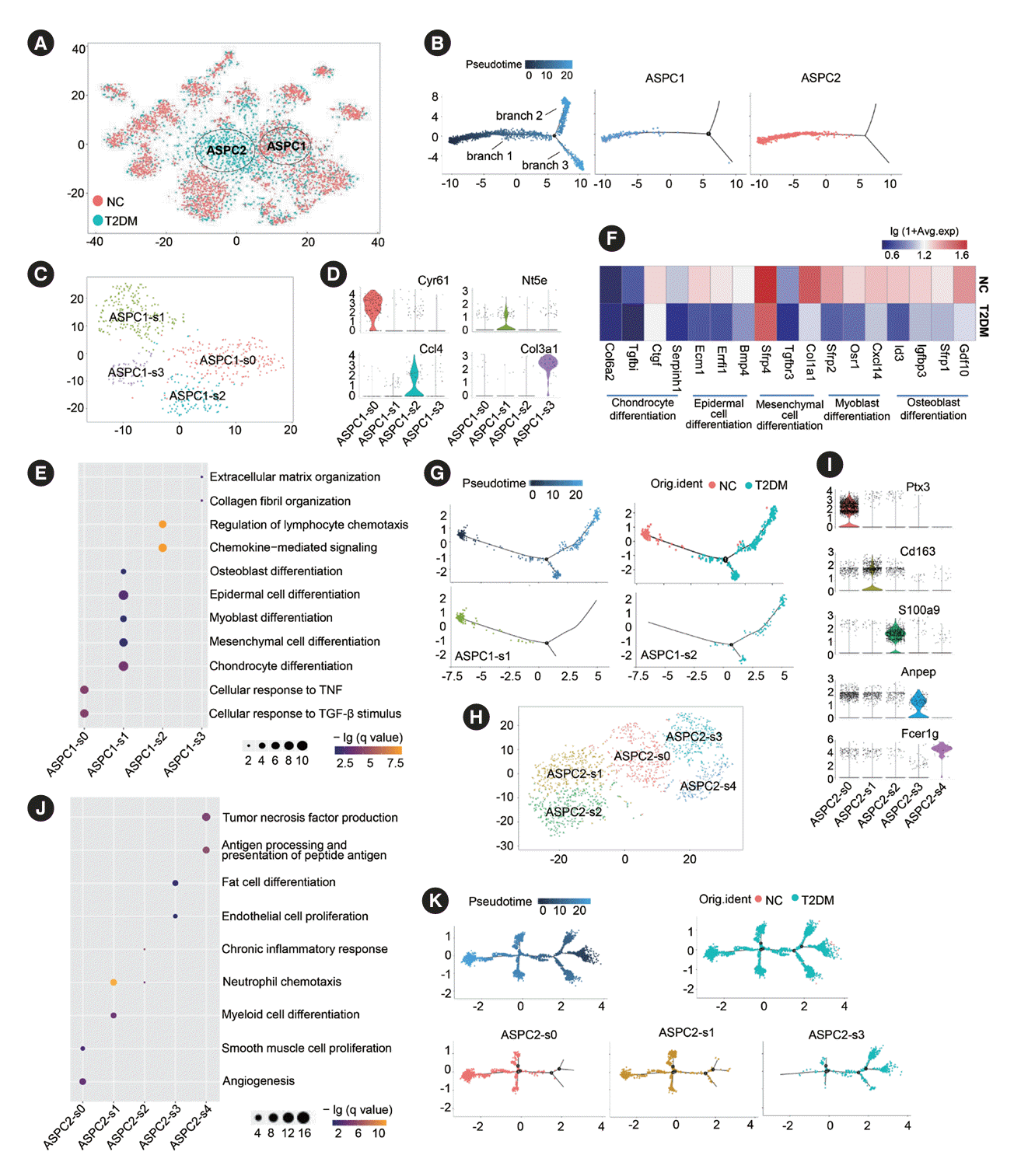
Fig. 4.
Subclusters and functions of endothelial cells (ECs) in brown adipose tissue (BAT). (A) The canonical correlation analysis (CCA; left panel) and t-distributed stochastic neighbor embedding (t-SNE; right panel) plots of EC subclusters in BAT. (B) Violin plots of relative expression levels of marker genes in each EC subcluster. (C) Functional enrichment analysis of each EC subcluster. (D) Representative immunofluorescence images of leucine rich repeat containing 8 VRAC subunit C (Lrrc8c)+cadherin 5 (Cdh5)+ECs in rat BAT. (E, F) Heatmap of relative expression levels (E) and functional enrichment analysis (F) of the significantly regulated genes in each EC subcluster in BAT of type 2 diabetes mellitus (T2DM) rats compared to those of normal rats (NC). Fisher’s exact test with Benjamini-Hochberg false discovery rate multiple-test correction was used for the statistical analysis in C and F. Mann-Whitney U test was performed for the statistical analysis in E. VEGF, vascular endothelial growth factor; SMC, smooth muscle cell; HDL, high density lipoprotein; DAPI, 4´,6-diamidino-2-phenylindole; TGF-β, transforming growth factor-β; ROS, reactive oxygen species; LDL, low density lipoprotein.
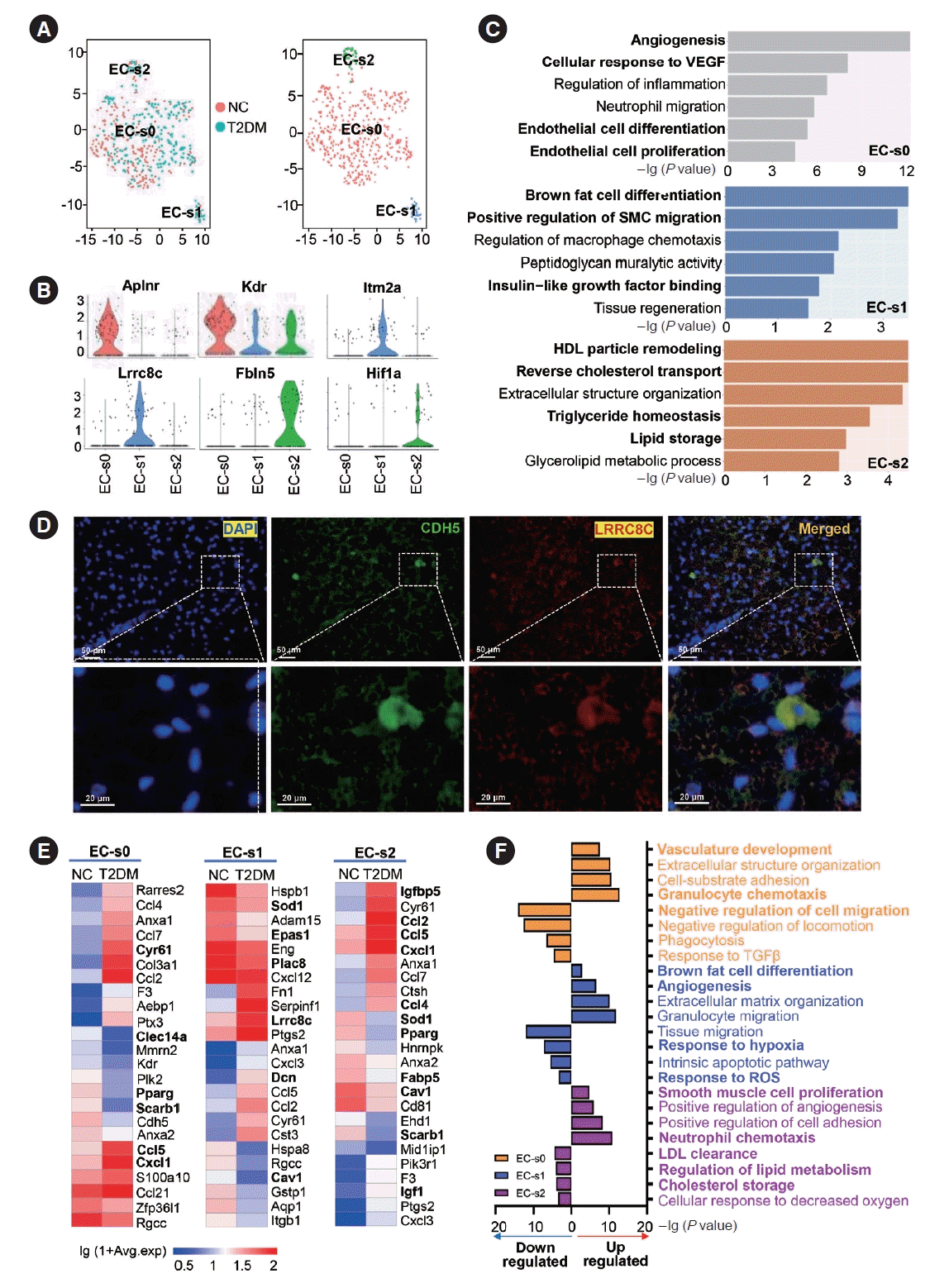
Fig. 5.
Subclusters and functions of macrophages (MACs) in brown adipose tissue (BAT). (A) The canonical correlation analysis (CCA; left panel) and t-distributed stochastic neighbor embedding (t-SNE; right panel) plots of MAC subclusters in BAT. (B) Violin plots of the relative expression levels of marker genes in each MAC subcluster. (C) Functional enrichment analysis of each MAC subcluster in BAT. (D) Proportion of each MAC subcluster in the total MACs in BAT of normal rats (NC) and type 2 diabetes mellitus (T2DM) rats. (E, F) Heatmap of relative expression levels (E) and functional enrichment analysis (F) of significantly regulated genes in each MAC subcluster of BAT in T2DM rats compared to those of NC rats. Fisher’s exact test with Benjamini-Hochberg false discovery rate multiple-test correction was used for the statistical analysis in C and F. Mann-Whitney U test was performed for the statistical analysis in E. ROS, reactive oxygen species; MHC, major histocompatibility complex; TNF, tumor necrosis factor; SMC, smooth muscle cell; TGF-β, transforming growth factor-β; PDGF, platelet derived growth factor.
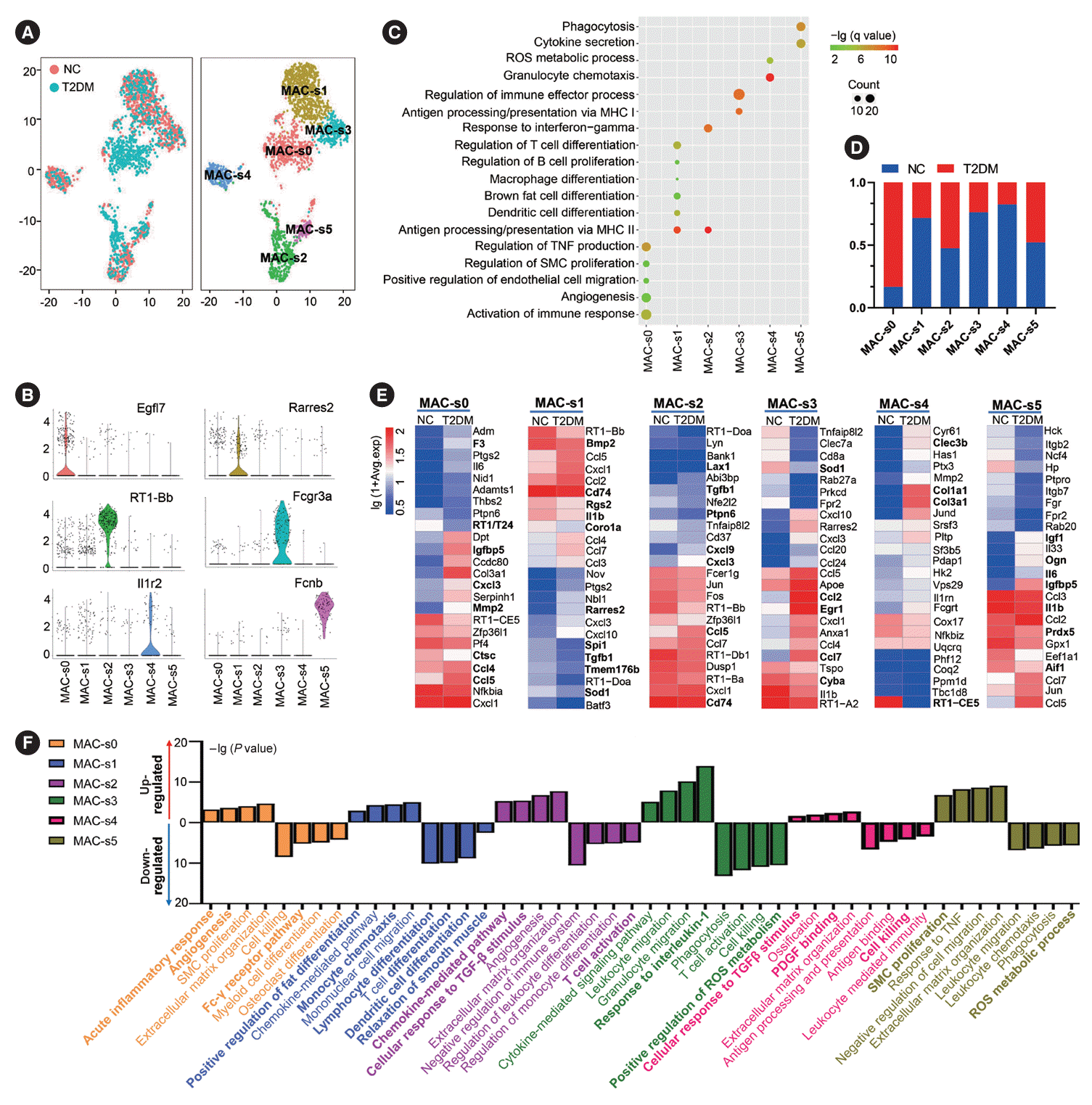
Fig. 6.
Retinoic acid receptor responder 2 (Rarres2)+macrophages (MACs) promote differentiation and function of brown adipocytes (BAs). (A) Representative immunofluorescence images of Rarres2+MACs in brown adipose tissue (BAT) of normal rats (NC). (B) Comparison of the percentage of Rarres2+MACs in white adipose tissue (WAT)-derived MACs and in BAT-derived MACs. (C) Comparison of the relative expression levels of Rarres2 mRNA in WAT-derived stromal vascular fraction (SVF), BA, and BAT-derived SVF. Fold change=The relative expression levels of each group compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH)/the mean of relative expression levels of WAT SVF compared to GAPDH. (D, E) Representative images of Oil Red O staining of brown adipocyte progenitor cells (BAPCs) with different treatments (D) and their quantitative comparison of the positive area of Oil Red O staining (E). Fold change=The positive area of Oil Red O staining of each group/the mean of positive area of Oil Red O staining of D0 BAPC. (F) Representative Western blot images of differentiation-related proteins and mitochondrial functional proteins in BAPCs with different treatments (left panel) and their quantitative comparison (right panel). (G) Quantitative comparison of the fluorescence intensity of mitochondria stained by Mitotracker Red in BAPCs with different treatments. Fold change=The fluorescence intensity of mitochondria stained of each group/the mean of fluorescence intensity of mitochondria stained of D0 BAPC. (H) Comparison of oxygen consumption rates (OCRs) at the plateau phase of BAPCs with different treatments. Fold change=The OCRs of each group/the mean of OCRs of D0 BAPC. (I) Representative Western blot images of phosphorylation levels of mammalian target of rapamycin (mTOR), AKT, and extracellular signal-regulated kinase (ERK) proteins, as well as expression levels of CCAAT enhancer binding protein beta (CEBPβ), CEBPα, and peroxisome proliferator-activated receptor γ (PPARγ) proteins in BAPCs with different treatments (left panel) and their quantitative comparison (right panel). D8 BAPC & Rarres2-MAC: BAPC were cocultured with Rarres2-MACs and differentiated on day 8; D8 BAPC & Rarres2+ MAC: BAPC were cocultured with Rarres2+MACs and differentiated on day 8. In F and I, fold change=The relative expression levels of each group compared to GAPDH/the mean of relative expression levels of D0 BAPC compared to GAPDH. Two-tailed Student’s t-test (B) and one-way analysis of variance (ANOVA) test with appropriate correction were used for statistical analysis (C, E-I). DAPI, 4´,6-diamidino-2-phenylindole; PRDM16, PR domain-containing 16; UCP1, uncoupling protein 1; COX7RP, cytochrome c oxidase subunit 7A2 like; TFAM, transcription factor A, mitochondrial. aP<0.05, bP<0.01, cP<0.001, dP<0.0001.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download