1. Neogi T. Clinical practice. Gout. N Engl J Med. 2011; 364:443–452. PMID:
21288096.
2. Li L, Yang C, Zhao Y, Zeng X, Liu F, Fu P. Is hyperuricemia an independent risk factor for new-onset chronic kidney disease?: a systematic review and meta-analysis based on observational cohort studies. BMC Nephrol. 2014; 15:122. PMID:
25064611.
3. Kuwabara M, Niwa K, Nishi Y, Mizuno A, Asano T, Masuda K, Komatsu I, Yamazoe M, Takahashi O, Hisatome I. Relationship between serum uric acid levels and hypertension among Japanese individuals not treated for hyperuricemia and hypertension. Hypertens Res. 2014; 37:785–789. PMID:
24671018.
4. Dehghan A, van Hoek M, Sijbrands EJ, Hofman A, Witteman JC. High serum uric acid as a novel risk factor for type 2 diabetes. Diabetes Care. 2008; 31:361–362. PMID:
17977935.
5. Zheng L, Zhu Y, Ma Y, Zhang H, Zhao H, Zhang Y, Yang Z, Liu Y. Relationship between hyperuricemia and the risk of cardiovascular events and chronic kidney disease in both the general population and hypertensive patients: a systematic review and meta-analysis. Int J Cardiol. 2024; 399:131779. PMID:
38218247.
6. Chen JH, Chuang SY, Chen HJ, Yeh WT, Pan WH. Serum uric acid level as an independent risk factor for all-cause, cardiovascular, and ischemic stroke mortality: a Chinese cohort study. Arthritis Rheum. 2009; 61:225–232. PMID:
19177541.
7. Trifirò G, Morabito P, Cavagna L, Ferrajolo C, Pecchioli S, Simonetti M, Bianchini E, Medea G, Cricelli C, Caputi AP, et al. Epidemiology of gout and hyperuricaemia in Italy during the years 2005-2009: a nationwide population-based study. Ann Rheum Dis. 2013; 72:694–700. PMID:
22736095.
8. Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007-2008. Arthritis Rheum. 2011; 63:3136–3141. PMID:
21800283.
9. Esche J, Krupp D, Mensink GB, Remer T. Dietary potential renal acid load is positively associated with serum uric acid and odds of hyperuricemia in the German adult population. J Nutr. 2018; 148:49–55. PMID:
29378039.
10. Singh G, Lingala B, Mithal A. Gout and hyperuricaemia in the USA: prevalence and trends. Rheumatology (Oxford). 2019; 58:2177–2180. PMID:
31168609.
11. Ting K, Gill TK, Keen H, Tucker GR, Hill CL. Prevalence and associations of gout and hyperuricaemia: results from an Australian population-based study. Intern Med J. 2016; 46:566–573. PMID:
26765205.
12. Zhang M, Zhu X, Wu J, Huang Z, Zhao Z, Zhang X, Xue Y, Wan W, Li C, Zhang W, et al. Prevalence of hyperuricemia among Chinese adults: findings from two nationally representative cross-sectional surveys in 2015–16 and 2018–19. Front Immunol. 2022; 12:791983. PMID:
35197964.
13. Jeong H, Chang YS, Jeon CH. Association between hyperuricemia and hearing impairment: results from the Korean National Health and Nutrition Examination Survey. Medicina (Kaunas). 2023; 59:1273. PMID:
37512084.
14. Lee CH, Sung NY. The prevalence and features of Korean gout patients using the National Health Insurance Corporation database. J Rheum Dis. 2011; 18:94–100.
15. Kim JW, Kwak SG, Lee H, Kim SK, Choe JY, Park SH. Prevalence and incidence of gout in Korea: data from the national health claims database 2007-2015. Rheumatol Int. 2017; 37:1499–1506. PMID:
28676911.
16. Williams PT. Effects of diet, physical activity and performance, and body weight on incident gout in ostensibly healthy, vigorously active men. Am J Clin Nutr. 2008; 87:1480–1487. PMID:
18469274.
17. Kaneko K, Aoyagi Y, Fukuuchi T, Inazawa K, Yamaoka N. Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia. Biol Pharm Bull. 2014; 37:709–721. PMID:
24553148.
18. Choi HK, Liu S, Curhan G. Intake of purine-rich foods, protein, and dairy products and relationship to serum levels of uric acid: the Third National Health and Nutrition Examination Survey. Arthritis Rheum. 2005; 52:283–289. PMID:
15641075.
19. Emmerson BT. The management of gout. N Engl J Med. 1996; 334:445–451. PMID:
8552148.
20. Schmidt JA, Crowe FL, Appleby PN, Key TJ, Travis RC. Serum uric acid concentrations in meat eaters, fish eaters, vegetarians and vegans: a cross-sectional analysis in the EPIC-Oxford cohort. PLoS One. 2013; 8:e56339. PMID:
23418557.
21. Miao Z, Li C, Chen Y, Zhao S, Wang Y, Wang Z, Chen X, Xu F, Wang F, Sun R, et al. Dietary and lifestyle changes associated with high prevalence of hyperuricemia and gout in the Shandong coastal cities of Eastern China. J Rheumatol. 2008; 35:1859–1864. PMID:
18634142.
22. Cho SK, Kim S, Chung JY, Jee SH. Discovery of URAT1 SNPs and association between serum uric acid levels and URAT1. BMJ Open. 2015; 5:e009360.
23. Jang WC, Nam YH, Park SM, Ahn YC, Park SH, Choe JY, Shin IH, Kim SK. T6092C polymorphism of
SLC22A12 gene is associated with serum uric acid concentrations in Korean male subjects. Clin Chim Acta. 2008; 398:140–144. PMID:
18824160.
24. Kolz M, Johnson T, Sanna S, Teumer A, Vitart V, Perola M, Mangino M, Albrecht E, Wallace C, Farrall M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009; 5:e1000504. PMID:
19503597.
25. Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A. 2009; 106:10338–10342. PMID:
19506252.
26. Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013; 45:145–154. PMID:
23263486.
27. Tin A, Marten J, Halperin Kuhns VL, Li Y, Wuttke M, Kirsten H, Sieber KB, Qiu C, Gorski M, Yu Z, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019; 51:1459–1474. PMID:
31578528.
28. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019; 51:584–591. PMID:
30926966.
29. Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019; 28:R133–R142. PMID:
31363735.
30. Nakatochi M, Kanai M, Nakayama A, Hishida A, Kawamura Y, Ichihara S, Akiyama M, Ikezaki H, Furusyo N, Shimizu S, et al. Genome-wide meta-analysis identifies multiple novel loci associated with serum uric acid levels in Japanese individuals. Commun Biol. 2019; 2:115. PMID:
30993211.
31. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020; 12:44. PMID:
32423490.
32. Choi SW, Mak TS, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020; 15:2759–2772. PMID:
32709988.
33. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018; 50:1219–1224. PMID:
30104762.
34. Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, Tyrer JP, Chen TH, Wang Q, Bolla MK, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. 2019; 104:21–34. PMID:
30554720.
35. Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018; 50:668–681. PMID:
29700475.
36. Janssens AC. Validity of polygenic risk scores: are we measuring what we think we are? Hum Mol Genet. 2019; 28:R143–R150. PMID:
31504522.
37. Kim Y, Han BG. KoGES group. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) consortium. Int J Epidemiol. 2017; 46:e20. PMID:
27085081.
38. Ahn Y, Kwon E, Shim JE, Park MK, Joo Y, Kimm K, Park C, Kim DH. Validation and reproducibility of food frequency questionnaire for Korean Genome Epidemiologic Study. Eur J Clin Nutr. 2007; 61:1435–1441. PMID:
17299477.
39. Lee KW, Woo HD, Cho MJ, Park JK, Kim SS. Identification of dietary patterns associated with incidence of hyperglycemia in middle-aged and older Korean adults. Nutrients. 2019; 11:1801. PMID:
31382699.
40. Jeong J, Lim K, Shin S. The association between meat intake and the risk of coronary heart disease in Korean men using the Framingham risk score: a prospective cohort study. Nutr Metab Cardiovasc Dis. 2023; 33:1158–1166. PMID:
36849318.
41. Korea Disease Control and Prevention Agency. Korean Genome and Epidemiology Study. Manual of Korean Genome and Epidemiology Study-Food Frequency Questionnaire [Internet]. Cheongju: Korea Disease Control and Prevention Agency;2019. cited 2024 July 24. Available from:
https://nih.go.kr/ko/main/contents.do?menuNo=300583.
42. Shin D, Lee KW. Dietary acid load is positively associated with the incidence of hyperuricemia in middle-aged and older Korean adults: findings from the Korean Genome and epidemiology study. Int J Environ Res Public Health. 2021; 18:10260. PMID:
34639563.
43. Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, Lu Y, Yoon K, Jang HM, Kim YK, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019; 9:1382. PMID:
30718733.
44. Ko B, Jin HS.
MACROD2 polymorphisms are associated with hypertension in Korean population. Korean J Clin Lab Sci. 2019; 51:57–63.
45. Park HJ, Kim SS, Jin HS. Genetic polymorphisms of SLC8A1 are associated with hypertension and left ventricular hypertrophy in the Korean population. Korean J Clin Lab Sci. 2019; 51:286–293.
46. Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, Yang J, Visscher PM, Wray NR. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019; 10:1891. PMID:
31015401.
47. Kim HR, Jin HS, Eom YB. Association between
MANBA gene variants and chronic kidney disease in a Korean population. J Clin Med. 2021; 10:2255. PMID:
34070965.
48. Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G, et al. Gene-obesogenic environment interactions in the UK Biobank study. Int J Epidemiol. 2017; 46:559–575. PMID:
28073954.
49. Lee WJ, Lim JE, Jung HU, Kang JO, Park T, Won S, Rhee SY, Kim MK, Kim YJ, Oh B. Analysis of the interaction between polygenic risk score and calorie intake in obesity in the Korean population. Lifestyle Genom. 2021; 14:20–29. PMID:
33302275.
50. Lin KC, Lin HY, Chou P. Community based epidemiological study on hyperuricemia and gout in Kin-Hu, Kinmen. J Rheumatol. 2000; 27:1045–1050. PMID:
10782835.
51. Byun SH, Yoo DM, Lee JW, Choi HG. Analyzing the association between hyperuricemia and periodontitis: a cross-sectional study using KoGES HEXA data. Int J Environ Res Public Health. 2020; 17:4777. PMID:
32630802.
52. Becker MA, Schumacher HR Jr, Wortmann RL, MacDonald PA, Eustace D, Palo WA, Streit J, Joseph-Ridge N. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N Engl J Med. 2005; 353:2450–2461. PMID:
16339094.
53. Nagahama K, Inoue T, Iseki K, Touma T, Kinjo K, Ohya Y, Takishita S. Hyperuricemia as a predictor of hypertension in a screened cohort in Okinawa, Japan. Hypertens Res. 2004; 27:835–841. PMID:
15824465.
54. Lee KW, Shin D. Concurrent presence of high serum uric acid and inflammation is associated with increased incidence of type 2 diabetes mellitus in Korean adult population. Sci Rep. 2022; 12:11000. PMID:
35768559.
55. Fu J, Shin S. The association of dietary patterns with incident chronic kidney disease and kidney function decline among middle-aged Korean adults: a cohort study. Epidemiol Health. 2023; 45:e2023037. PMID:
37311642.
56. Moon ME, Jung DH, Heo SJ, Park B, Lee YJ. Oxidative balance score and new-onset type 2 diabetes mellitus in Korean adults without non-alcoholic fatty liver disease: Korean Genome and Epidemiology Study-Health Examinees (KoGES-HEXA) cohort. Antioxidants. 2024; 13:107. PMID:
38247531.
57. Azadbakht L, Esmaillzadeh A. Red meat intake is associated with metabolic syndrome and the plasma C-reactive protein concentration in women. J Nutr. 2009; 139:335–339. PMID:
19074209.
58. Lee SA, Kwon SO, Song M, Choi JY, Shin A, Shu XO, Zheng W, Lee JK, Kang D. The association of serum high-sensitivity C-reactive protein level with the risk of site-specific cancer mortality: the Health Examinees (HEXA) study cohort. Am J Epidemiol. 2022; 191:2002–2013. PMID:
35916370.
59. Cho GJ, Kim J, Kim JY, Han SW, Lee SB, Oh MJ, Kim SJ, Shin JE. Adverse pregnancy outcomes and maternal chronic diseases in the future: a cross-sectional study using KoGES-HEXA data. J Clin Med. 2022; 11:1457. PMID:
35268548.
60. Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, Mollace V. Regulation of uric acid metabolism and excretion. Int J Cardiol. 2016; 213:8–14. PMID:
26316329.
61. Mandal AK, Mount DB. The molecular physiology of uric acid homeostasis. Annu Rev Physiol. 2015; 77:323–345. PMID:
25422986.
62. Rivera-Paredez B, Macías-Kauffer L, Fernandez-Lopez JC, Villalobos-Comparán M, Martinez-Aguilar MM, de la Cruz-Montoya A, Ramírez-Salazar EG, Villamil-Ramírez H, Quiterio M, Ramírez-Palacios P, et al. Influence of genetic and non-genetic risk factors for serum uric acid levels and hyperuricemia in Mexicans. Nutrients. 2019; 11:1336. PMID:
31207883.
63. Abreu E, Fonseca MJ, Santos AC. Association between hyperuricemia and insulin resistance. Acta Med Port. 2011; 24(Suppl 2):565–574.
64. Rule AD, de Andrade M, Matsumoto M, Mosley TH, Kardia S, Turner ST. Association between SLC2A9 transporter gene variants and uric acid phenotypes in African American and white families. Rheumatology (Oxford). 2011; 50:871–878. PMID:
21186168.
65. Brandstätter A, Kiechl S, Kollerits B, Hunt SC, Heid IM, Coassin S, Willeit J, Adams TD, Illig T, Hopkins PN, et al. Sex-specific association of the putative fructose transporter
SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care. 2008; 31:1662–1667. PMID:
18487473.
66. So A, Thorens B. Uric acid transport and disease. J Clin Invest. 2010; 120:1791–1799. PMID:
20516647.
67. Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008; 372:1953–1961. PMID:
18834626.
68. Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, Hajat C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008; 82:139–149. PMID:
18179892.
69. Vitart V, Rudan I, Hayward C, Gray NK, Floyd J, Palmer CN, Knott SA, Kolcic I, Polasek O, Graessler J, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008; 40:437–442. PMID:
18327257.
70. Döring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, Fischer G, Henke K, Klopp N, Kronenberg F, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008; 40:430–436. PMID:
18327256.
71. McArdle PF, Parsa A, Chang YP, Weir MR, O’Connell JR, Mitchell BD, Shuldiner AR. Association of a common nonsynonymous variant in
GLUT9 with serum uric acid levels in old order amish. Arthritis Rheum. 2008; 58:2874–2881. PMID:
18759275.
72. Guan M, Zhou D, Ma W, Chen Y, Zhang J, Zou H. Association of an intronic SNP of
SLC2A9 gene with serum uric acid levels in the Chinese male Han population by high-resolution melting method. Clin Rheumatol. 2011; 30:29–35. PMID:
20972595.
73. Lyngdoh T, Bochud M, Glaus J, Castelao E, Waeber G, Vollenweider P, Preisig M. Associations of serum uric acid and
SLC2A9 variant with depressive and anxiety disorders: a population-based study. PLoS One. 2013; 8:e76336. PMID:
24204615.
74. Sun X, Jiang F, Zhang R, Tang SS, Chen M, Peng DF, Yan J, Wang T, Wang SY, Bao YQ, et al. Serum uric acid levels are associated with polymorphisms in the
SLC2A9,
SF1, and
GCKR genes in a Chinese population. Acta Pharmacol Sin. 2014; 35:1421–1427. PMID:
25283508.
75. Hu X, Rong S, Wang Q, Sun T, Bao W, Chen L, Liu L. Association between plasma uric acid and insulin resistance in type 2 diabetes: a Mendelian randomization analysis. Diabetes Res Clin Pract. 2021; 171:108542. PMID:
33227361.
76. Sun X, Zhang R, Jiang F, Tang S, Chen M, Peng D, Yan J, Wang T, Wang S, Bao Y, et al. Common variants related to serum uric acid concentrations are associated with glucose metabolism and insulin secretion in a Chinese population. PLoS One. 2015; 10:e0116714. PMID:
25617895.
77. Zhao J, Guo S, Schrodi SJ, He D. Trends in the contribution of genetic susceptibility loci to hyperuricemia and gout and associated novel mechanisms. Front Cell Dev Biol. 2022; 10:937855. PMID:
35813212.
78. Wong K, Briddon SJ, Holliday ND, Kerr ID. Plasma membrane dynamics and tetrameric organisation of ABCG2 transporters in mammalian cells revealed by single particle imaging techniques. Biochim Biophys Acta. 2016; 1863:19–29. PMID:
26453803.
79. Li R, Miao L, Qin L, Xiang Y, Zhang X, Peng H, Mailamuguli , Sun Y, Yao H. A meta-analysis of the associations between the Q141K and Q126X ABCG2 gene variants and gout risk. Int J Clin Exp Pathol. 2015; 8:9812–9823. PMID:
26617691.
80. Yu KH, Chang PY, Chang SC, Wu-Chou YH, Wu LA, Chen DP, Lo FS, Lu JJ. A comprehensive analysis of the association of common variants of ABCG2 with gout. Sci Rep. 2017; 7:9988. PMID:
28855613.
81. Lee S, Yang HK, Lee HJ, Park DJ, Kong SH, Park SK. Cross-phenotype association analysis of gastric cancer: in-silico functional annotation based on the disease-gene network. Gastric Cancer. 2023; 26:517–527. PMID:
36995485.
82. Wang J, Liu S, Wang B, Miao Z, Han L, Chu N, Zhang K, Meng D, Li C, Ma X. Association between gout and polymorphisms in
GCKR in male Han Chinese. Hum Genet. 2012; 131:1261–1265. PMID:
22395765.
83. Urano W, Taniguchi A, Inoue E, Sekita C, Ichikawa N, Koseki Y, Kamatani N, Yamanaka H. Effect of genetic polymorphisms on development of gout. J Rheumatol. 2013; 40:1374–1378. PMID:
23729800.
84. Wang L, Ma Q, Yao H, He LJ, Fang BB, Cai W, Zhang B, Wang ZQ, Su YX, Du GL, et al. Association of GCKR rs780094 polymorphism with circulating lipid levels in type 2 diabetes and hyperuricemia in Uygur Chinese. Int J Clin Exp Pathol. 2018; 11:4684–4694. PMID:
31949869.
85. Ho LJ, Lu CH, Su RY, Lin FH, Su SC, Kuo FC, Chu NF, Hung YJ, Liu JS, Hsieh CH. Association between glucokinase regulator gene polymorphisms and serum uric acid levels in Taiwanese adolescents. Sci Rep. 2022; 12:5519. PMID:
35365700.
86. Rasheed H, Stamp LK, Dalbeth N, Merriman TR. Interaction of the
GCKR and
A1CF loci with alcohol consumption to influence the risk of gout. Arthritis Res Ther. 2017; 19:161. PMID:
28679452.
87. Christensen EI, Birn H. Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol. 2002; 3:256–266. PMID:
11994745.
88. Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010; 42:210–215. PMID:
20139978.
89. Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018; 50:390–400. PMID:
29403010.
90. Rasheed H, Phipps-Green A, Topless R, Hollis-Moffatt JE, Hindmarsh JH, Franklin C, Dalbeth N, Jones PB, White DH, Stamp LK, et al. Association of the lipoprotein receptor-related protein 2 gene with gout and non-additive interaction with alcohol consumption. Arthritis Res Ther. 2013; 15:R177. PMID:
24286387.
91. Lan B, Chen P, Jiri M, He N, Feng T, Liu K, Jin T, Kang L.
WDR1 and
CLNK gene polymorphisms correlate with serum glucose and high-density lipoprotein levels in Tibetan gout patients. Rheumatol Int. 2016; 36:405–412. PMID:
26438387.
92. Cervero P, Himmel M, Krüger M, Linder S. Proteomic analysis of podosome fractions from macrophages reveals similarities to spreading initiation centres. Eur J Cell Biol. 2012; 91:908–922. PMID:
22721921.
93. Kile BT, Panopoulos AD, Stirzaker RA, Hacking DF, Tahtamouni LH, Willson TA, Mielke LA, Henley KJ, Zhang JG, Wicks IP, et al. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood. 2007; 110:2371–2380. PMID:
17515402.
94. Wu JN, Koretzky GA. The SLP-76 family of adapter proteins. Semin Immunol. 2004; 16:379–393. PMID:
15541653.
95. Yu J, Riou C, Davidson D, Minhas R, Robson JD, Julius M, Arnold R, Kiefer F, Veillette A. Synergistic regulation of immunoreceptor signaling by SLP-76-related adaptor Clnk and serine/threonine protein kinase HPK-1. Mol Cell Biol. 2001; 21:6102–6112. PMID:
11509653.
96. Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010; 3:523–530. PMID:
20884846.
97. Hong M, Park JW, Yang PS, Hwang I, Kim TH, Yu HT, Uhm JS, Joung B, Lee MH, Jee SH, et al. A Mendelian randomization analysis: the causal association between serum uric acid and atrial fibrillation. Eur J Clin Invest. 2020; 50:e13300. PMID:
32474920.
98. Chiba T, Matsuo H, Kawamura Y, Nagamori S, Nishiyama T, Wei L, Nakayama A, Nakamura T, Sakiyama M, Takada T, et al. NPT1/SLC17A1 is a renal urate exporter in humans and its common gain-of-function variant decreases the risk of renal underexcretion gout. Arthritis Rheumatol. 2015; 67:281–287. PMID:
25252215.
99. Torres RJ, de Miguel E, Bailén R, Banegas JR, Puig JG. Tubular urate transporter gene polymorphisms differentiate patients with gout who have normal and decreased urinary uric acid excretion. J Rheumatol. 2014; 41:1863–1870. PMID:
25128519.
100. Park JS, Kim Y, Kang J. Genome-wide meta-analysis revealed several genetic loci associated with serum uric acid levels in Korean population: an analysis of Korea Biobank data. J Hum Genet. 2022; 67:231–237. PMID:
34719683.
101. Dong Z, Zhou J, Jiang S, Li Y, Zhao D, Yang C, Ma Y, Wang Y, He H, Ji H, et al. Effects of multiple genetic loci on the pathogenesis from serum urate to gout. Sci Rep. 2017; 7:43614. PMID:
28252667.
102. Chen BD, Chen XC, Pan S, Yang YN, He CH, Liu F, Ma X, Gai MT, Ma YT. TT genotype of rs2941484 in the human
HNF4G gene is associated with hyperuricemia in Chinese Han men. Oncotarget. 2017; 8:26918–26926. PMID:
28460474.
103. Wisely GB, Miller AB, Davis RG, Thornquest AD Jr, Johnson R, Spitzer T, Sefler A, Shearer B, Moore JT, Miller AB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002; 10:1225–1234. PMID:
12220494.
104. Kawaguchi M, Nakayama A, Aoyagi Y, Nakamura T, Shimizu S, Kawamura Y, Takao M, Tamura T, Hishida A, Nagayoshi M, et al. Both variants of
A1CF and
BAZ1B genes are associated with gout susceptibility: a replication study and meta-analysis in a Japanese population. Hum Cell. 2021; 34:293–299. PMID:
33517564.
105. Leask MP, Merriman TR. The genetic basis of urate control and gout: insights into molecular pathogenesis from follow-up study of genome-wide association study loci. Best Pract Res Clin Rheumatol. 2021; 35:101721. PMID:
34732286.
106. Gottier Nwafor J, Nowik M, Anzai N, Endou H, Wagner CA. Metabolic acidosis alters expression of Slc22 transporters in mouse kidney. Kidney Blood Press Res. 2020; 45:263–274. PMID:
32062662.
107. Jamshidi N, Nigam KB, Nigam SK. Loss of the kidney urate transporter, Urat1, leads to disrupted redox homeostasis in mice. Antioxidants. 2023; 12:780. PMID:
36979028.
108. Sugihara S, Hisatome I, Kuwabara M, Niwa K, Maharani N, Kato M, Ogino K, Hamada T, Ninomiya H, Higashi Y, et al. Depletion of uric acid due to SLC22A12 (URAT1) loss-of-function mutation causes endothelial dysfunction in hypouricemia. Circ J. 2015; 79:1125–1132. PMID:
25739858.
109. Shima Y, Teruya K, Ohta H. Association between intronic SNP in urate-anion exchanger gene,
SLC22A12, and serum uric acid levels in Japanese. Life Sci. 2006; 79:2234–2237. PMID:
16920156.
110. Matsuo H, Yamamoto K, Nakaoka H, Nakayama A, Sakiyama M, Chiba T, Takahashi A, Nakamura T, Nakashima H, Takada Y, et al. Genome-wide association study of clinically defined gout identifies multiple risk loci and its association with clinical subtypes. Ann Rheum Dis. 2016; 75:652–659. PMID:
25646370.
111. Yang Y, Lin C, Zheng Q, Zhang L, Li Y, Huang Q, Wu T, Zhao Z, Li L, Luo J, et al. L-carnitine attenuated hyperuricemia-associated left ventricular remodeling through ameliorating cardiomyocytic lipid deposition. Front Pharmacol. 2023; 14:1016633. PMID:
36817129.
112. Cuevas-Delgado P, Miguel V, Rupérez FJ, Lamas S, Barbas C. Impact of renal tubular
Cpt1a overexpression on the kidney metabolome in the folic acid-induced fibrosis mouse model. Front Mol Biosci. 2023; 10:1161036. PMID:
37377862.
113. Estiverne C, Mandal AK, Mount DB. Molecular pathophysiology of uric acid homeostasis. Semin Nephrol. 2020; 40:535–549. PMID:
33678309.
114. Narang RK, Vincent Z, Phipps-Green A, Stamp LK, Merriman TR, Dalbeth N. Population-specific factors associated with fractional excretion of uric acid. Arthritis Res Ther. 2019; 21:234. PMID:
31718705.
115. Merriman TR. An update on the genetic architecture of hyperuricemia and gout. Arthritis Res Ther. 2015; 17:98. PMID:
25889045.
116. Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci U S A. 1981; 78:6858–6862. PMID:
6947260.
117. Leask M, Dowdle A, Salvesen H, Topless R, Fadason T, Wei W, Schierding W, Marsman J, Antony J, O’Sullivan JM, et al. Functional urate-associated genetic variants influence expression of lincRNAs
LINC01229 and
MAFTRR
. Front Genet. 2019; 9:733. PMID:
30719032.
118. Merriman T, Phipps-Green A, Topless R, Merriman M, Franklin C, Jones G, van Rij A, Montgomery G, Chapman B, White D, et al. Association analysis of 18 recently discovered serum urate loci with gout. Ann Rheum Dis. 2014; 73:353.
119. Phipps-Green AJ, Merriman ME, Topless R, Altaf S, Montgomery GW, Franklin C, Jones GT, van Rij AM, White D, Stamp LK, et al. Twenty-eight loci that influence serum urate levels: analysis of association with gout. Ann Rheum Dis. 2016; 75:124–130. PMID:
25187157.
120. Li C, Li Z, Liu S, Wang C, Han L, Cui L, Zhou J, Zou H, Liu Z, Chen J, et al. Genome-wide association analysis identifies three new risk loci for gout arthritis in Han Chinese. Nat Commun. 2015; 6:7041. PMID:
25967671.
121. Sakiyama M, Matsuo H, Nakaoka H, Kawamura Y, Kawaguchi M, Higashino T, Nakayama A, Akashi A, Ueyama J, Kondo T, et al. Common variant of
BCAS3 is associated with gout risk in Japanese population: the first replication study after gout GWAS in Han Chinese. BMC Med Genet. 2018; 19:96. PMID:
29879923.
122. Lourida I, Hannon E, Littlejohns TJ, Langa KM, Hyppönen E, Kuzma E, Llewellyn DJ. Association of lifestyle and genetic risk with incidence of dementia. JAMA. 2019; 322:430–437. PMID:
31302669.
123. Rutten-Jacobs LC, Larsson SC, Malik R, Rannikmäe K; MEGASTROKE Consortium; International Stroke Genetics Consortium, Sudlow CL, Dichgans M, Markus HS, Traylor M. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. BMJ. 2018; 363:k4168. PMID:
30355576.
124. Patel AP, Wang M, Ruan Y, Koyama S, Clarke SL, Yang X, Tcheandjieu C, Agrawal S, Fahed AC, Ellinor PT, et al. A multi-ancestry polygenic risk score improves risk prediction for coronary artery disease. Nat Med. 2023; 29:1793–1803. PMID:
37414900.
125. Said MA, Verweij N, van der Harst P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK Biobank study. JAMA Cardiol. 2018; 3:693–702. PMID:
29955826.
126. Arthur RS, Wang T, Xue X, Kamensky V, Rohan TE. Genetic factors, adherence to healthy lifestyle behavior, and risk of invasive breast cancer among women in the UK Biobank. J Natl Cancer Inst. 2020; 112:893–901. PMID:
31899501.
127. Xin J, Du M, Gu D, Jiang K, Wang M, Jin M, Hu Y, Ben S, Chen S, Shao W, et al. Risk assessment for colorectal cancer via polygenic risk score and lifestyle exposure: a large-scale association study of East Asian and European populations. Genome Med. 2023; 15:4. PMID:
36694225.
128. Zhang T, Gu Y, Meng G, Zhang Q, Liu L, Wu H, Zhang S, Wang X, Zhang J, Sun S, et al. Genetic risk, adherence to a healthy lifestyle, and hyperuricemia: the TCLSIH cohort study. Am J Med. 2023; 136:476–483.e5. PMID:
36708795.
129. Hak AE, Choi HK. Menopause, postmenopausal hormone use and serum uric acid levels in US women--the Third National Health and Nutrition Examination Survey. Arthritis Res Ther. 2008; 10:R116. PMID:
18822120.
130. Xiong Z, Zhu C, Qian X, Zhu J, Wu Z, Chen L. Serum uric acid is associated with dietary and lifestyle factors in elderly women in suburban Guangzhou in Guangdong province of south China. J Nutr Health Aging. 2013; 17:30–34. PMID:
23299375.
131. Li R, Yu K, Li C. Dietary factors and risk of gout and hyperuricemia: a meta-analysis and systematic review. Asia Pac J Clin Nutr. 2018; 27:1344–1356. PMID:
30485934.
132. Zhang T, Gan S, Ye M, Meng G, Zhang Q, Liu L, Wu H, Gu Y, Zhang S, Wang Y, et al. Association between consumption of ultra-processed foods and hyperuricemia: TCLSIH prospective cohort study. Nutr Metab Cardiovasc Dis. 2021; 31:1993–2003. PMID:
34119375.
133. Rai SK, Fung TT, Lu N, Keller SF, Curhan GC, Choi HK. The Dietary Approaches to Stop Hypertension (DASH) diet, Western diet, and risk of gout in men: prospective cohort study. BMJ. 2017; 357:j1794. PMID:
28487277.
134. Zhang M, Chang H, Gao Y, Wang X, Xu W, Liu D, Li G, Huang G. Major dietary patterns and risk of asymptomatic hyperuricemia in Chinese adults. J Nutr Sci Vitaminol (Tokyo). 2012; 58:339–345. PMID:
23327969.
135. Xia Y, Xiang Q, Gu Y, Jia S, Zhang Q, Liu L, Meng G, Wu H, Bao X, Yu B, et al. A dietary pattern rich in animal organ, seafood and processed meat products is associated with newly diagnosed hyperuricaemia in Chinese adults: a propensity score-matched case-control study. Br J Nutr. 2018; 119:1177–1184. PMID:
29759111.
136. Lin K, McCormick N, Yokose C, Joshi AD, Lu N, Curhan GC, Merriman TR, Saag KG, Ridker PM, Buring JE, et al. Interactions between genetic risk and diet influencing risk of incident female gout: discovery and replication analysis of four prospective cohorts. Arthritis Rheumatol. 2023; 75:1028–1038. PMID:
36512683.
137. Roman YM. Moving the needle in gout management: the role of culture, diet, genetics, and personalized patient are practices. Nutrients. 2022; 14:3590. PMID:
36079846.
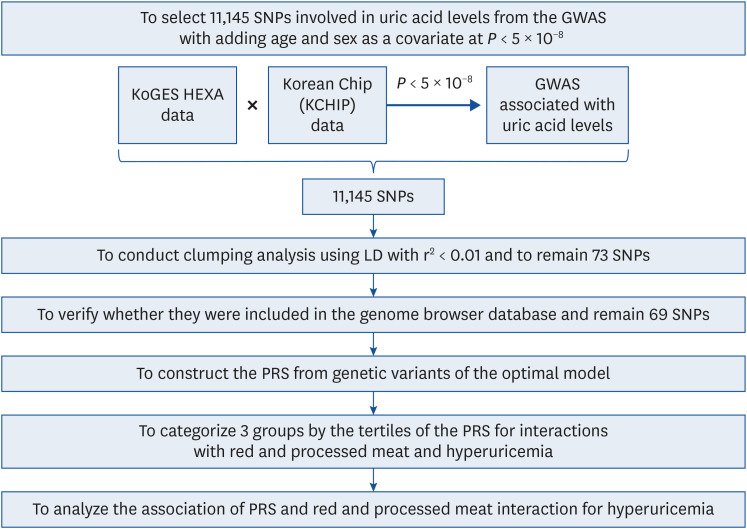
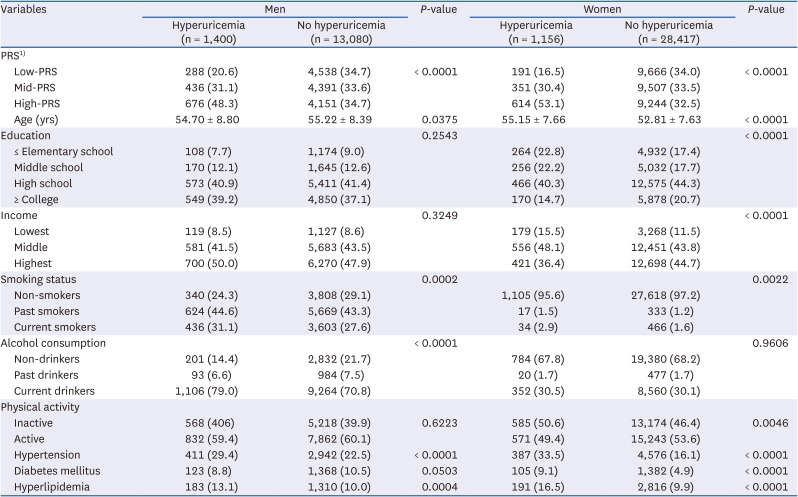
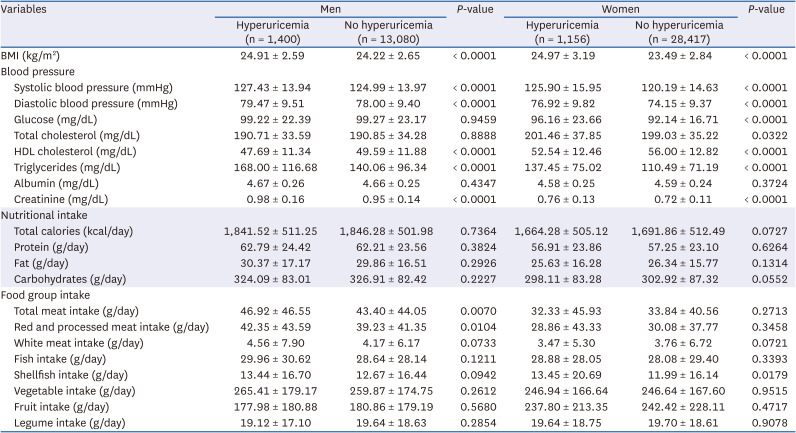
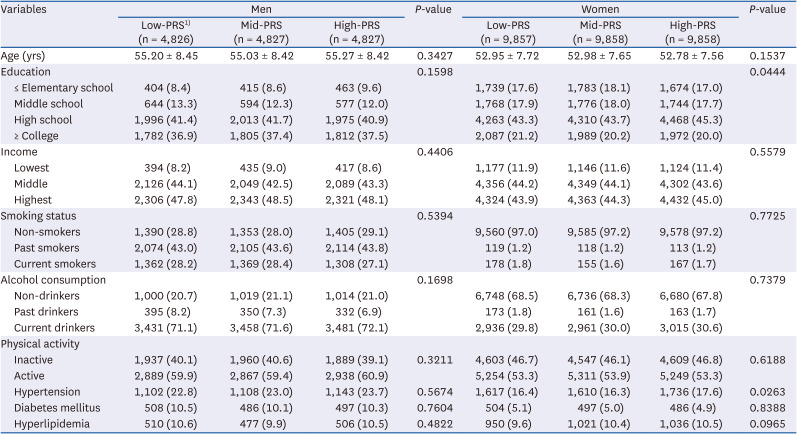




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download