Abstract
Background and Objectives
The colonic epithelial layer is a complex structure consisting of multiple cell types that regulate various aspects of colonic physiology, yet the mechanisms underlying epithelial cell differentiation during development remain unclear. Organoids have emerged as a promising model for investigating organogenesis, but achieving organ-like cell configurations within colonic organoids is challenging. Here, we investigated the biological significance of peripheral neurons in the formation of colonic organoids.
Methods and Results
Colonic organoids were co-cultured with human embryonic stem cell (hESC)-derived peripheral neurons, resulting in the morphological maturation of columnar epithelial cells, as well as the presence of enterochromaffin cells. Substance P released from immature peripheral neurons played a critical role in the development of colonic epithelial cells. These findings highlight the vital role of inter-organ interactions in organoid development and provide insights into colonic epithelial cell differentiation mechanisms.
The colonic epithelial layer is a complex and dynamic structure that consists of multiple cell types, including goblet cells, enterocytes, Paneth cells, enteroendocrine cells, and intestinal stem cells (1). Each of these cells plays a crucial role in regulating various aspects of colonic physiology, such as mucosal formation, colonic absorption, colonic motility, immunity, and regeneration (2). Although these cells are derived from colonic stem cells (3), the mechanisms underlying their diversification during development are not yet fully understood.
Organoids, which are three-dimensional structures that recapitulate the process of organ formation in vitro, have emerged as a promising research model for investigating organogenesis and understanding the mechanisms underlying cell diversification (4). However, achieving organ-like cell configurations within colonic organoids remains a significant challenge (5). Since various types of cells arise as the end product of the developmental process, simulating the developmental process during the organoid establishment is recognized as a necessary condition. Such simulations require appropriate intrinsic and extrinsic signaling stimuli, and recent research has shown that interactions between cells, tissues, or organs also play an important role. One of the most significant examples of inter-organ interaction is the connection between nerves and target organs (6-8). Peripheral nerves not only accept and deliver environmental information and commands but also contribute to the maintenance of structural and functional homeostasis and the development of the target organ to which they connect (9-11). In the case of the large intestine, peripheral nerve connections are observed from the early stages of organ formation (12), however, the precise role of the nervous system in colonic development remains unclear.
The aim of this study was to evaluate the biological significance of peripheral neuron contributions in the formation of colonic organoids. We co-cultured colonic organoids with human embryonic stem cell (hESC)-derived peripheral neurons following induction of colonic specification through temporarily restricted stimulation of bone morphogenetic protein (BMP). Our investigation revealed that substance P (SP) spontaneously released from immature peripheral neurons plays a critical role in the development of colonic epithelial cells, specifically colon enterochromaffin cells. Our findings highlight the essential role of inter-organ interactions in organoid development and provide insights into the mechanisms underlying colonic epithelial cell differentiation.
The hESC line WA09 (H9) was obtained from WiCell. The cells were maintained in Essential 8 medium (Thermo Fisher, USA) on iMatrix-511 coating material (Matrixome, Japan) under a 5% CO2 condition in a humidified incu-bator. To prevent any unintended effects of genetic mutations, hESCs between passages 30 to 40 were collected using either Versene solution (Thermo Fisher, USA) or Accutase (Sigma Aldrich, USA) for further experiments. All experiments involving hESCs were conducted under institutional IRB regulation KHSIRB-20-489 approved by Kyung Hee University (13).
To generate colonic organoids, we modified previously reported protocols (14-16). Briefly, 2.4 million hESCs were plated in a 24-well plate and allowed to reach 70% confluency as day 0. From day 0 to day 3, definitive endoderm (DE) was induced using RPMI 1640 medium (Welgene, Korea) containing 2% fetal bovine serum (FBS) (Welgene, Korea), 3 uM CHIR99021 (Sigma Aldrich, USA), and 100 ng/ml Activin A (Peprotech, USA). From day 4 to day 7, hindgut cells were differentiated using DMEM/F-12 medium (Welgene, Korea) containing 2% FBS, 1% B27 (Thermo Fisher, USA), 3 uM CHIR99021, and 500 ng/ml fibroblast growth factor 4 (FGF4) (Peprotech, USA), and then the resulting cell aggregates were embedded in Geltrex (Thermo Fisher, USA) after detachment using dispase (Thermo Fisher, USA). From day 8, DMEM/F-12 medium containing 1% B-27, 3 uM CHIR99021, 100 nM LDN193189 (STEMGENT, USA), and 100 ng/ml epidermal growth factor (EGF) (Peprotech, USA) was applied continuously, and from day 14 to day 20, 20 ng/ml BMP4 (Peprotech, USA) was temporarily added to the same medium composition to generate colonic organoids.
To differentiate NCSCs from PHOX2B::eGFP reporter hESCs, colonies were dissociated into single cells using Accutase (Sigma Aldrich) and plated on Geltrex (Thermo Fisher, USA), allowing them to reach 70%∼80% confluence following previous protocols (17). Differentiation into NCSCs was achieved by gradually changing Essential 8 medium to Neurobasal medium (Thermo Fisher, USA) supplemented with B27 and N-2 (Thermo Fisher, USA), containing 500 nM LDN193189 and 10 μM SB431542 for the first three days, followed by the addition of 3 μM CHIR99021 for an additional eight days. FACS sorting was used to isolate NCSCs expressing surface antigen p75 nerve growth factor receptor (NGFR) and human natural killer 1 (HNK-1). Enteric NCSCs were differentiated using a previously reported protocol as a control (18). After reaching 70%∼80% confluence on Geltrex, cells were treated with 500 nM LDN193189 and 10 μM SB431542 in gradually changing Neurobasal medium from day 0 to day 3. From day 4 to day 6, cells were treated with 3 μM CHIR99021, SB431542, 10 μM DAPT (Cayman Chemi-cal, USA), and 0.2 μM PD173074 (Sigma Aldrich, USA). From day 7 to day 12, cells were treated with 100 ng/ml sonic hedgehog (SHH) (Peprotech, USA) and 1 μM purmorphamine (PMP) (Sigma Aldrich). From day 10 to 14, cells were treated with 10 μg/ml BMP4. On day 14, PHOX2B::eGFP-expressing cells were sorted by FACS. Peripheral neurons were further differentiated by adding 10 nM nerve growth factor (NGF) (Peprotech, USA), 20 nM brain derived neurotrophic factor (BDNF) (Peprotech, USA), 20 nM glial cell derived neurotrophic factor (GDNF) (Peprotech, USA), 100 μM dibutyryl cyclic adenosine monophosphate (dbcAMP) (Sigma Aldrich, USA), and 100 μM sodium L-ascorbate (Sigma Aldrich, USA) to the culture medium, and cells were cultured for an additional seven weeks (13).
FACS-sorted NGFR and HNK1 double-positive NCSCs (250,000 cells/well) were cultured on Matrigel-coated plates in Neurobasal medium supplemented with B27 and N-2, 20 ng/ml bFGF, and 20 ng/ml EGF for 24 hours. After washing the colonic organoids embedded in Matrigel with cold PBS, they were separated from the matrix using Cell Recovery Solution (Corning, USA). Approximately 50 organoids per well were allowed to naturally settle onto the NCSCs. The co-culture medium for colonic organoids and NCSCs was a 1:1 mixture of Neurobasal medium containing B27 and N-2 supplements and DMEM/F-12 medium. Half of the medium was changed every other day with fresh medium. To establish co-culture without direct contact between colonic organoids and NCSCs, NGFR and HNK1 double-positive NCSCs and colonic organoids were cultured separately on Matrigel-coated plates for 24 hours in B27 and N-2-supplemented Neurobasal medium containing 20 ng/ml EGF and 20 ng/ml bFGF or DMEM/F-12 medium. A transwell system was used, where the colonic organoids were cultured in the lower chamber and the NCSCs in the upper chamber, with the two chambers separated by a 0.4 μm pore membrane. The culture medium was a 1:1 mixture of Neurobasal medium and DMEM/F-12, and half of the medium was changed every other day with a fresh one.
For flow cytometry analysis, the cells were mechanically dissociated after exposure to Accutase and treated with 40 μg/ml DNase I (Roche Applied Science, Germa-ny) for 20 minutes at 37℃. Next, the cells were incubated with appropriate antibodies for 1 hour at 4℃, washed twice with Phosphate Buffered Saline (DPBS), and treated with 7-AAD (BD Pharmingen, USA) to eliminate dead cells, following the manufacturer’s instructions. The cells were analyzed using an FC500 (Beckman Coulter, USA) or isolated using FACS.
Cells and cryosectioned organoid tissues were fixed with 4% paraformaldehyde and permeabilized with PBS containing 0.5% Triton X-100. After blocking with 5% normal goat serum (Jackson Laboratory, USA) in PBST, the cells were incubated overnight at 4℃ with primary antibodies. The primary antibodies were diluted in PBS with 0.1% normal serum. The cells were then incubated with secondary antibodies at a 1:500 dilution in PBS for 1 hour at room temperature. Finally, the slides were mounted using a mounting medium with DAPI (Vector labs, USA) and imaged using fluorescence microscopy (Olympus Cor-poration, Japan). To eliminate any background fluoresce-nce, negative controls without primary antibodies were included in the experiment. Antibodies used in the research are listed in Table 1.
Total RNA was extracted using Trizol (Invitrogen), RNA concentration was measured spectrophotometrically using a Nanodrop (DeNvix Inc., USA). Subsequently, 2 μg of total RNA was reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, USA). The quantitative PCR was performed on the StepOne Plus Real-Time PCR system (Applied Biosystems, USA) using AccuPower GreenSrat qPCR PreMix (Bioneer, Korea). Relative mRNA levels were normalized to those of 18S for each gene. The primer sequences are listed in Table 1.
Proteins were separated by SDS-PAGE and transferred onto nitrocellulose (NC) membranes (Bio-Rad, USA) for immunoblotting. Immunoblotting was performed with indicated primary antibodies (Table 1) and relevant secondary antibodies (Thermo Fisher, USA). Immunoblot bands were visualized with chemiluminescence (Pierce, USA) and developed manually using X-ray film (Kodak, USA).
All data were analyzed using GraphPad Prism 9.0 software (GraphPad, USA). The data were presented as mean± standard deviation (SD) of at least three independent biological experiments. Morphological images representative of at least three biologically independent experiments with similar results were captured. The statistical significance was evaluated using paired t-test, unpaired t-tests, one-way ANOVA, or two-way ANOVA as appropriate. A p-value less than 0.05 was considered statistically significant. The bands’ densitometric analysis for immunoblotting was conducted using FIJI software (Image J, USA). FlowJo 10.0 software (Treestar, USA) was utilized to analyze the FACS data. The illustrations were generated using images obtained from biorender.com.
We developed a stepwise protocol for generating colon organoids by modifying previously reported protocols for colonic organoids, which either stimulated BMP signals (14) or did not (15) (Fig. 1A). The differentiation process was characterized by a decrease in the pluripotency markers OCT4 and NANOG (Fig. 1B). By day 3 of differen-tiation towards DE, cells expressed both mRNA and protein for SOX17 and FOXA2 (Fig. 1C and 1D), with over 70% of cells expressing each protein (Fig. 1E and 1F). With additional differentiation, cells on day 7 expressed mRNA and protein for CDX1 and KLF5 (Fig. 1G and 1H) and differentiated into hindgut cells that formed cell clusters (Fig. 1I). The differentiation rate at this stage was similar to that of DE differentiation, with over 70% of cells expressing each protein (Fig. 1J). During the process of inducing the development of colonic organoids by embedding hindgut cells that formed clusters into an artificial ECM matrix in clump form (Fig. 1K), the addition of BMP4 in the latter stages of embedding significantly increased the expression of hindgut hox genes, HOXD12, HOXA13, HOXB13, and HOXD13, which are responsible for posterior development, compared to hox gene families responsible for development of the foregut or midgut (Fig. 1L). The application of BMP4 to the colonic organoids also resulted in an increase in mRNA and protein expression of special AT-rich sequence-binding protein 2 (SATB2), a colon-restricted chromatin factor of the large intestine in the posterior region (Fig. 1M and 1N) (19). Additionally, a cell cluster expressing the SATB2 protein was identified in the inner center of the colonic organoid (Fig. 1O).
After long-term culture, colonic organoids embedded in the ECM matrix exhibited an increase in volume as the ECM matrix was gradually removed (Fig. 2A). At week 6, Mucin proteins (MUC), detectable by MUC2, MUC3, and MUC4 antibodies, accumulated within the organoids (Fig. 2B), indicating the differentiation of epithelial cells, including goblet cells. Flow cytometry analysis of EpCAM-expressing cells revealed that approximately 8% of the cells were epithelial (Fig. 2C). Immunostaining confirmed the formation of an epithelial layer expressing EpCAM, CD133, or Villin on the outer layer of the organoids (Fig. 2D). To identify the various cell types within the epithelial layer, markers representing each cell type were examined. MUC3, MUC4, Villin, defensin alpha 5 (DEFA5), and leucine rich repeat containing G protein-coupled receptor 5 (LGR5) expression were detected (Fig. 2E), indicating the presence of goblet cells, enterocytes, Paneth cells, and colonic stem cells. However, chromogranin A (CHGA) expression, a marker for enteroendocrine cells, was not detected, suggesting that enteroendocrine cells were not present in the epithelial layer of the organoids (Fig. 2E). To assess the functionality of the epithelial cells based on their integrity, we treated the organoids with forskolin and performed a swelling assay based on CF transmembrane conductance regulator (CFTR) activation (20). As a result, we confirmed that the organoids swelled in response to forskolin treatment, indicating the presence of functional epithelial cells (Fig. 2F).
To investigate the reciprocal interaction between immature embryonic colonic tissue and the peripheral nervous system, we co-cultured colon organoids with NCSCs that were differentiated from PHOX2B::eGFP reporter hESCs (18) using a conventional method (21) (Fig. 3A). Within 7 days of co-culture, NCSCs showed morphological characteristics of neuronal differentiation (Fig. 3B), and about 5% of NCSCs expressed eGFP, indicating the expression of PHOX2B (Fig. 3C). FACS sorting was used to isolate NCSCs expressing PHOX2B after co-culture with colon organoids, which showed increased expression of PHOX2B and the enteric neuronal marker N-myc downstream-regulated gene 4 (NDRG4), compared to NCSCs not co-cultured with colon organoids (Fig. 3D). Meanwhile, colon organoids isolated after co-culture showed an enrichment of mRNA expression for enterochromaffin cell markers LIM homeobox transcription factor 1 alpha (LMX1A) and CHGA (Fig. 3E). To determine whether the reciprocal effects were due to direct contact or unidirectional influence, we attempted co-culture using transwells (Fig. 3F). The proportion of PHOX2B-expressing cells induced by co-culture with colon organoids decreased compared to NCSCs in direct contact with colon organoids, but still maintained an increased proportion compared to NCSCs that were not co-cultured (Fig. 3G). The isolated PHOX2B-expressing cells were verified for their expre-ssion (Fig. 3H). In contrast, the expression of NDRG4 and LMX1A in colon organoids co-cultured with NCSCs via transwells did not increase (Fig. 3I), unlike colon organoids in direct contact with NCSCs.
After co-culturing NCSCs and colon organoids directly for 10 days, differentiated neuronal cells expressing neuron-specific class III beta-tubulin (TUJ1) contained serotonin (Fig. 3J, lower panel), and the epithelial cells of colon organoids labeled with eGFP surrounded by the neuronal cells exhibited typical columnar morphology of colon epithelial cells (Fig. 3J). However, co-culture with NCSCs did not increase the number of epithelial cells expressing EpCAM (Fig. 3K). Instead, the proportion of Ki-67-ex-pressing cells, which are proliferating cells, decreased within the colon organoids (Fig. 3L). When EpCAM-expressing cells were separated from the colon organoids after co-culture with NCSCs and re-plated, epithelial cells expressing EpCAM showed low efficiency for attachment and survival rates while non-epithelial cells that did not express EpCAM exhibited high survival and proliferation rates (Fig. 3M). After 10 days of re-cultivation, most rapidly proliferating cells from EpCAM-expressing cells expressed SOX9, an epithelial stem cell marker in the colon (22) (Fig. 3N).
To investigate the influence of developing neurons on colonic organoids, we examined the time points during co-culture of NCSCs and colonic organoids at which LMX1A and NDRG4 changed (Fig. 4A). Between days 4 and 6 of co-culture, we observed an increasing trend in both NDRG4 in NCSC-derived neurons and LMX1A in colonic orga-noids. This coincided with a significant difference in TUJ1 expression, indicating neuronal differentiation of NCSCs (Fig. 4B). To further evaluate the impact of developing neurons on colonic organoids, we measured the quantity of a signaling molecule, substance P, which is spontaneously secreted by immature peptidergic neurons during early differentiation. We used ELISA to measure substance P levels in the co-culture medium on days 4 and 6. The result revealed a significant increase in substance P secretion on day 6 compared to day 4 in the NCSC-derived neurons co-cultured with colonic organoids (Fig. 4C). While the level of substance P was lower than that produced by immature neurons spontaneously secreting substance P alone, it was much higher than that of matured neurons or NCSCs not co-cultured with colonic organoids (Fig. 4C). Upon direct administration of substance P to colonic organoids, we observed a significant decrease in the expression of DEFA5 protein, while the expression of CHGA protein increased (Fig. 4D). Interestingly, we found that cells expressing CHGA were scattered throughout the organoid instead of being limited to the epithelial layer (Fig. 4E). Additionally, the epithelial cells outside of the organoid maintained their columnar morphology, and we were able to confirm the presence of enteroendo-crine cells detectable by 5-hydroxytryptamine (5-HT) within the organoid (Fig. 4F) (23). Notably, the expression of neurokinin 1 receptor (NK1R), which is known as a receptor for substance P (24), increased in colonic organoids co-cultured with NCSC-derived neurons or treated with Substance P (Fig. 4G). To investigate whether the interaction between peripheral neurons and colonic organoids due to a conventional mechanism of substance P via NK1R, we utilized NK1R antagonists for inhibition. How-ever, the increase in CHGA protein level was not significantly attenuated (Fig. 4H). The findings suggest that substance P plays an important role in regulating the expression of NK1R and cells expressing CHGA in colonic organoids, but it does not selectively target the cells expressing NK1R in a conventional state.
In this study, we developed a comprehensive protocol for generating colon organoids that closely resemble key features of the developing colon. The protocol involves inducing differentiation of cells towards definitive endoderm, followed by embedding hindgut cells in an artificial extracellular matrix (ECM), and stimulating BMP4 to initiate colonic differentiation. Our hypothesis was that BMP4 plays a crucial role in determining the orientation of cells, particularly the epithelial lining, instead of in overall gut tube formation since it is expressed symmetrically only in the gut tube and is not detectable in the mesentery (25-27). Our findings revealed that adding BMP4 instead of previously used BMP2 (15) during the later stage of the culture period induced the formation of epithelial cells with posterior gut tube specification (19, 28). These organoids exhibited functional epithelial cells such as goblet cells, enterocytes, Paneth cells, and colonic stem cells, as evidenced by the presence of markers such as MUC3, MUC4, Villin, DEFA5, and LGR5 (29, 30). Additionally, the organoids responded to forskolin treatment, indicating the pre-sence of functional epithelial cells (20). Despite these promising results, enteroendocrine cells were not detected in the epithelial layer of the organoids. Thus, further optimization was required to promote their differentiation.
To investigate the interaction between colonic organoids and the peripheral nervous system, we co-cultured colonic organoids with NCSCs. We observed that NCSCs differentiated into enteric NCSCs expressing PHOX2B and neurons expressing TUJ1 (18), while colonic organoids showed abundant mRNA expression for the enterochromaffin cell markers LMX1A and CHGA. This suggests that the interaction between colon organoids and NCSC-derived neurons promotes enteric chromaffin cell differentiation, further implying the potential role of the enteric nervous system in gut development (12). Interestingly, co-cultured colon organoids and NCSCs spontaneously differentiated into peripheral neurons expressing PHOX2B without any specific stimulation, modeling the behavior of migrating NCSCs naturally committing to enteric fate when approaching the developing colon along the dorso-ventral track in vivo (21, 31). Moreover, substance P, which was secreted from early differentiated neurons in co-culture, has been found to promote intestinal epithelial differen-tiation. This observation suggests that immature early differentiated neurons possess the ability to spontaneously release neuropeptides nonsynaptically and contribute to tissue formation during development (32-35).
However, our study has limitations of oversimplifying the complex interaction between developing immature gut and differentiating peripheral neurons. The enteric nervous system in the colon is composed of various types of neurons and enteric glial cells (36), forming complex neural networks in the submucosal and myenteric plexuses (37). To clarify the impact of interactions between immature organoids and developing neurons on the functional maturation of the gut epithelium and the functional specification of neural cells, it is necessary to differentiate the various types of neural cell constituents and conduct further investigations into the precise mechanisms underlying nonsynaptic transmission of substance P.
Despite certain limitations, our research sheds light on the crucial role of immature neurons in the process of development. Our study indicates that even though the colon requires fewer enterochromaffin cells compared to the small intestine, peripheral nerve innervation is essential for the presence of the limited number of enterochro-maffin cells in the colon. Our results also suggest that irregular neural activation or dysfunctional nerve cells that regulate neuropeptides and neurotransmitters such as substance P may lead to abnormal increases in enterochroma-ffin cells in the rectum of irritable bowel syndrome (IBS) patients (38-41). Currently, there is a dearth of research models for neurological disorders like IBS, and our colonic organoids innervated by neurons may prove invaluable for studying relevant mechanisms and developing therapeutic interventions to regulate neuropeptides and neuro-transmitters. Moreover, considering the role of the microbiome in gut health, further studies may explore the interaction between colon organoids and the microbiota in the gut. Understanding the interplay between the microbiota and the nervous system may help unravel the complex mechanisms underlying the development of gastrointestinal disorders and facilitate the development of more effective treatments (42, 43).
Overall, our findings have far-reaching implications for the fields of developmental biology and tissue engineering. The stepwise protocol we used to generate colon organoids can be optimized further to facilitate the differentiation of other cell types in the colonic epithelial cells. Additio-nally, investigating the interaction between colon organoids and the peripheral nervous system can deepen our understanding of the mechanisms that regulate gut development, function, and diseases. Future studies may lead to the development of novel therapeutic interventions for gastrointestinal disorders and enhance our knowledge of the complexities of organ development and function.
Notes
Author Contributions
Y.H.C., I.Y.C., and C.E.S.: performing experiments, data analysis, assembly, interpretation, and writing manuscript; C.P., S.K.L., and J.H.K.: performing experiments and data analysis; S.H.S.: performing experiments; S.L. and J.H.P: study design; Y.J.K.: conception, study design, performing experiments, data analysis, assembly, interpretation, and writing a manuscript.
References
1. Frazer LC, Good M. 2022; Intestinal epithelium in early life. Mucosal Immunol. 15:1181–1187. DOI: 10.1038/s41385-022-00579-8. PMID: 36380094. PMCID: PMC10329854.

2. Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. 2017; Mend Your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 4:33–46. DOI: 10.1016/j.jcmgh.2017.03.007. PMID: 28560287. PMCID: PMC5439240.
3. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. 2007; Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. DOI: 10.1038/nature06196. PMID: 17934449.

4. Lancaster MA, Knoblich JA. 2014; Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 345:1247125. DOI: 10.1126/science.1247125. PMID: 25035496.

5. Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, Sugimoto S, Sato T. 2018; Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 23:787–793.e6. DOI: 10.1016/j.stem.2018.11.016. PMID: 30526881.

6. Najjar SA, Davis BM, Albers KM. 2020; Epithelial-neuronal communication in the colon: implications for visceral pain. Trends Neurosci. 43:170–181. DOI: 10.1016/j.tins.2019.12.007. PMID: 31983457. PMCID: PMC7047572.

7. Walsh KT, Zemper AE. 2019; The enteric nervous system for epithelial researchers: basic anatomy, techniques, and interactions with the epithelium. Cell Mol Gastroenterol Hepatol. 8:369–378. DOI: 10.1016/j.jcmgh.2019.05.003. PMID: 31108231. PMCID: PMC6718943.

8. Ten Hove AS, Seppen J, de Jonge WJ. 2021; Neuronal innervation of the intestinal crypt. Am J Physiol Gastrointest Liver Physiol. 320:G193–G205. DOI: 10.1152/ajpgi.00239.2020. PMID: 33296267.

9. Zhang W, Lyu M, Bessman NJ, Xie Z, Arifuzzaman M, Yano H, Parkhurst CN, Chu C, Zhou L, Putzel GG, Li TT, Jin WB, Zhou J, Hu H, Tsou AM, Guo CJ, Artis D. JRI Live Cell Bank. 2022; Gut-innervating nociceptors regulate the intestinal microbiota to promote tissue protection. Cell. 185:4170–4189.e20. DOI: 10.1016/j.cell.2022.09.008. PMID: 36240781.

10. Yang D, Jacobson A, Meerschaert KA, Sifakis JJ, Wu M, Chen X, Yang T, Zhou Y, Anekal PV, Rucker RA, Sharma D, Sontheimer-Phelps A, Wu GS, Deng L, Anderson MD, Choi S, Neel D, Lee N, Kasper DL, Jabri B, Huh JR, Johansson M, Thiagarajah JR, Riesenfeld SJ, Chiu IM. 2022; Nociceptor neurons direct goblet cells via a CGRP-RAMP1 axis to drive mucus production and gut barrier protection. Cell. 185:4190–4205.e25. DOI: 10.1016/j.cell.2022.09.024. PMID: 36243004.

11. Worthington JJ, Reimann F, Gribble FM. 2018; Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 11:3–20. DOI: 10.1038/mi.2017.73. PMID: 28853441.

12. Nagy N, Goldstein AM. 2017; Enteric nervous system development: a crest cell's journey from neural tube to colon. Semin Cell Dev Biol. 66:94–106. DOI: 10.1016/j.semcdb.2017.01.006. PMID: 28087321. PMCID: PMC5474363.

13. Lyoo KS, Kim HM, Lee B, Che YH, Kim SJ, Song D, Hwang W, Lee S, Park JH, Na W, Yun SP, Kim YJ. 2022; Direct neuronal infection of SARS-CoV-2 reveals cellular and molecular pathology of chemosensory impairment of COVID-19 patients. Emerg Microbes Infect. 11:406–411. DOI: 10.1080/22221751.2021.2024095. PMID: 34962444. PMCID: PMC8803065. PMID: f5c15fb6b0404ecea7026b9d0ae175ef.

14. Crespo M, Vilar E, Tsai SY, Chang K, Amin S, Srinivasan T, Zhang T, Pipalia NH, Chen HJ, Witherspoon M, Gordillo M, Xiang JZ, Maxfield FR, Lipkin S, Evans T, Chen S. 2017; Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med. 23:878–884. DOI: 10.1038/nm.4355. PMID: 28628110. PMCID: PMC6055224.

15. Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, Hudson JR, Howell JC, Chatuvedi P, Spence JR, Shannon JM, Zorn AM, Helmrath MA, Wells JM. 2017; Differe-ntiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. 21:51–64.e6. DOI: 10.1016/j.stem.2017.05.020. PMID: 28648364. PMCID: PMC5531599.

16. Kwon O, Yu WD, Son YS, Jung KB, Lee H, Son MY. 2022; Generation of highly expandable intestinal spheroids composed of stem cells. Int J Stem Cells. 15:104–111. DOI: 10.15283/ijsc21209. PMID: 35220296. PMCID: PMC8889332.

17. Mukherjee-Clavin B, Mi R, Kern B, Choi IY, Lim H, Oh Y, Lannon B, Kim KJ, Bell S, Hur JK, Hwang W, Che YH, Habib O, Baloh RH, Eggan K, Brandacher G, Hoke A, Studer L, Kim YJ, Lee G. 2019; Comparison of three congruent patient-specific cell types for the modelling of a human genetic Schwann-cell disorder. Nat Biomed Eng. 3:571–582. DOI: 10.1038/s41551-019-0381-8. PMID: 30962586. PMCID: PMC6612317.

18. Oh Y, Cho GS, Li Z, Hong I, Zhu R, Kim MJ, Kim YJ, Tampakakis E, Tung L, Huganir R, Dong X, Kwon C, Lee G. 2016; Functional coupling with cardiac muscle promotes maturation of hPSC-derived sympathetic neurons. Cell Stem Cell. 19:95–106. DOI: 10.1016/j.stem.2016.05.002. PMID: 27320040. PMCID: PMC4996639.

19. Gu W, Wang H, Huang X, Kraiczy J, Singh PNP, Ng C, Dagdeviren S, Houghton S, Pellon-Cardenas O, Lan Y, Nie Y, Zhang J, Banerjee KK, Onufer EJ, Warner BW, Spence J, Scherl E, Rafii S, Lee RT, Verzi MP, Redmond D, Longman R, Helin K, Shivdasani RA, Zhou Q. 2022; SATB2 preserves colon stem cell identity and mediates ileum-colon conversion via enhancer remodeling. Cell Stem Cell. 29:101–115.e10. DOI: 10.1016/j.stem.2021.09.004. PMID: 34582804. PMCID: PMC8741647.

20. Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NW, Bijvelds MJ, Scholte BJ, Nieuwenhuis EE, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. 2013; A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 19:939–945. DOI: 10.1038/nm.3201. PMID: 23727931.

21. Kim YJ, Lim H, Li Z, Oh Y, Kovlyagina I, Choi IY, Dong X, Lee G. 2014; Generation of multipotent induced neural crest by direct reprogramming of human postnatal fibroblasts with a single transcription factor. Cell Stem Cell. 15:497–506. DOI: 10.1016/j.stem.2014.07.013. PMID: 25158936.

22. Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD, Magness ST. 2012; Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am J Physiol Gastrointest Liver Physiol. 302:G10–G20. DOI: 10.1152/ajpgi.00277.2011. PMID: 21995959. PMCID: PMC3345960.
23. Linan-Rico A, Ochoa-Cortes F, Beyder A, Soghomonyan S, Zuleta-Alarcon A, Coppola V, Christofi FL. 2016; Mechanosensory signaling in enterochromaffin cells and 5-HT release: potential implications for gut inflammation. Front Neurosci. 10:564. DOI: 10.3389/fnins.2016.00564. PMID: 28066160. PMCID: PMC5165017. PMID: 1bf89b535afe480d81ba16b4085a369e.

24. Goode T, O'Connor T, Hopkins A, Moriarty D, O'Sullivan GC, Collins JK, O'Donoghue D, Baird AW, O'Connell J, Shanahan F. 2003; Neurokinin-1 receptor (NK-1R) expression is induced in human colonic epithelial cells by proinflam-matory cytokines and mediates proliferation in response to substance P. J Cell Physiol. 197:30–41. DOI: 10.1002/jcp.10234. PMID: 12942538.

25. Nerurkar NL, Mahadevan L, Tabin CJ. 2017; BMP signaling controls buckling forces to modulate looping morphogenesis of the gut. Proc Natl Acad Sci U S A. 114:2277–2282. DOI: 10.1073/pnas.1700307114. PMID: 28193855. PMCID: PMC5338480.

26. Castillo D, Puig S, Iglesias M, Seoane A, de Bolós C, Munitiz V, Parrilla P, Comerma L, Poulsom R, Krishnadath KK, Grande L, Pera M. 2012; Activation of the BMP4 pathway and early expression of CDX2 characterize non-specialized columnar metaplasia in a human model of Barrett's esophagus. J Gastrointest Surg. 16:227–237. discussion 237DOI: 10.1007/s11605-011-1758-5. PMID: 22076569.

27. Zhou G, Sun YG, Wang HB, Wang WQ, Wang XW, Fang DC. 2009; Acid and bile salt up-regulate BMP4 expression in human esophageal epithelium cells. Scand J Gastroenterol. 44:926–932. DOI: 10.1080/00365520902998661. PMID: 19488929.

28. Moore-Scott BA, Opoka R, Lin SC, Kordich JJ, Wells JM. 2007; Identification of molecular markers that are expressed in discrete anterior-posterior domains of the endoderm from the gastrula stage to mid-gestation. Dev Dyn. 236:1997–2003. DOI: 10.1002/dvdy.21204. PMID: 17576135.

29. Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, Ashley N, Johnson E, Hublitz P, Bao L, Lukomska J, Andev RS, Björklund E, Kessler BM, Fischer R, Goldin R, Koohy H, Simmons A. 2019; Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 567:49–55. DOI: 10.1038/s41586-019-0992-y. PMID: 30814735.

30. Dharshika C, Gulbransen BD. 2023; Enteric neuromics: how high-throughput "Omics" deepens our understanding of enteric nervous system genetic architecture. Cell Mol Gastroe-nterol Hepatol. 15:487–504. DOI: 10.1016/j.jcmgh.2022.10.019. PMID: 36368612. PMCID: PMC9792566.

31. Uesaka T, Young HM, Pachnis V, Enomoto H. 2016; Development of the intrinsic and extrinsic innervation of the gut. Dev Biol. 417:158–167. DOI: 10.1016/j.ydbio.2016.04.016. PMID: 27112528.

32. McMahon D. 1974; Chemical messengers in development: a hypothesis. Science. 185:1012–1021. DOI: 10.1126/science.185.4156.1012. PMID: 4152810.

33. LoTurco JJ, Owens DF, Heath MJ, Davis MB, Kriegstein AR. 1995; GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 15:1287–1298. DOI: 10.1016/0896-6273(95)90008-X. PMID: 8845153.

34. Demarque M, Represa A, Becq H, Khalilov I, Ben-Ari Y, Aniksztejn L. 2002; Paracrine intercellular communication by a Ca2+− and SNARE-independent release of GABA and glutamate prior to synapse formation. Neuron. 36:1051–1061. DOI: 10.1016/S0896-6273(02)01053-X. PMID: 12495621.

35. Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. 2007; GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 87:1215–1284. DOI: 10.1152/physrev.00017.2006. PMID: 17928584.

36. Yun W, Kim YJ, Lee G. 2022; Direct conversion to achieve glial cell fates: oligodendrocytes and Schwann cells. Int J Stem Cells. 15:14–25. DOI: 10.15283/ijsc22008. PMID: 35220289. PMCID: PMC8889328.

37. Wang H, Foong JPP, Harris NL, Bornstein JC. 2022; Enteric neuroimmune interactions coordinate intestinal responses in health and disease. Mucosal Immunol. 15:27–39. DOI: 10.1038/s41385-021-00443-1. PMID: 34471248. PMCID: PMC8732275.

38. Dunlop SP, Jenkins D, Neal KR, Spiller RC. 2003; Relative importance of enterochromaffin cell hyperplasia, anxiety, and depression in postinfectious IBS. Gastroenterology. 125:1651–1659. DOI: 10.1053/j.gastro.2003.09.028. PMID: 14724817.

39. Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, Pasquinelli G, Morselli-Labate AM, Grady EF, Bunnett NW, Collins SM, Corinaldesi R. 2004; Acti-vated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastro-enterology. 126:693–702. DOI: 10.1053/j.gastro.2003.11.055. PMID: 14988823.

40. Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA. 2006; Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology. 130:34–43. DOI: 10.1053/j.gastro.2005.09.031. PMID: 16401466.

41. Barbara G, Wang B, Stanghellini V, de Giorgio R, Cremon C, Di Nardo G, Trevisani M, Campi B, Geppetti P, Tonini M, Bunnett NW, Grundy D, Corinaldesi R. 2007; Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 132:26–37. DOI: 10.1053/j.gastro.2006.11.039. PMID: 17241857.

42. Park NY, Koh A. 2022; From the dish to the real world: modeling interactions between the gut and microorganisms in gut organoids by tailoring the gut milieu. Int J Stem Cells. 15:70–84. DOI: 10.15283/ijsc21243. PMID: 35220293. PMCID: PMC8889331.

43. Lee H, Son MY. 2021; Current challenges associated with the use of human induced pluripotent stem cell-derived organoids in regenerative medicine. Int J Stem Cells. 14:9–20. DOI: 10.15283/ijsc20140. PMID: 33632980. PMCID: PMC7904522.

Fig. 1
Stepwise protocol for generating colonic organoids. (A) Schematic diagram showing the modification of previously reported protocols for colonic organoids, with or without bone morphogenetic protein (BMP) signaling stimulation. (B) Decrease in pluripotency markers OCT4 and NANOG during differentiation process. (C) Expression level of mRNA for SOX17 and FOXA2 on day 3 of differentiation towards definitive endoderm (DE). (D) Immunoblot images of protein expression for SOX17 and FOXA2 on the differentiation stage of DE. GAPDH was used as an internal control. (E) Cells expressing a protein for SOX17 and FOXA2 on the DE stage. (F) Counting results for SOX17 and FOXA2 expressing cells from (E). (G) Expression level of mRNA for CDX1 and KLF5 on day 7 of differentiation towards hindgut (HG) cells. (H) Immunoblot images of protein expression for CDX1 and KLF5 on the differentiation stage of hindgut. GAPDH was used as an internal control. (I) Cells expressing a protein for CDX1 and KLF5 on the hindgut stage. (J) Counting results for CDX1 and KLF5 expressing cells from (I). (K) Schematic diagram showing the embedding process of hindgut cells that formed clusters into an artificial ECM matrix in clump form. (L) BMP4 stimulation during the embedding stage increases expression of hindgut hox genes responsible for posterior development. (M and N) Increase in mRNA and protein expression of colon-restricted chromatin factor SATB2 with BMP4 application. (O) Identification of a cell cluster expressing the SATB2 protein in the inner center of the colonic organoid. Data expressed as mean±SD (n=9, biological repeat). p-values calculated by one-way ANOVA (B, L, M) or unpaired t-test (C, F, G, and J). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant. All scale bars=50 μm. DE: definitive endoderm, HG: hindgut, CO: colonic organoid.
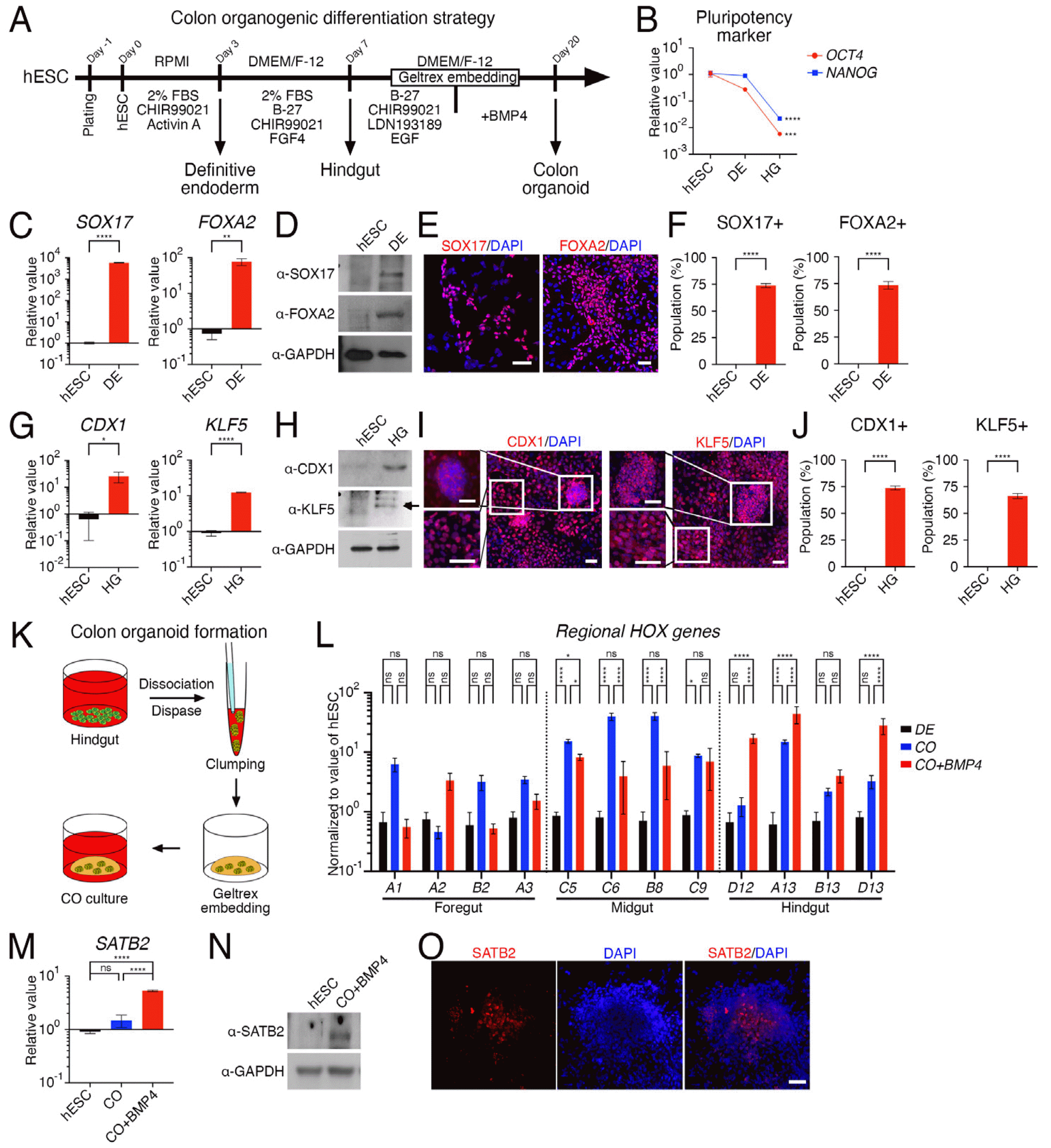
Fig. 2
Characterization of long-term cultured colonic organoids embedded in an ECM matrix. (A) Morphological changes of colonic organoids during long-term culture. (B) Detection of Mucin proteins (MUC) within the organoids at week 6 using MUC2, MUC3, and MUC4 antibodies. (C) Representative FACS plot for EpCAM-expressing cells of colonic organoids. (D) Immunostaining confirmed the formation of an epithelial layer expressing EpCAM, CD133, or Villin on the outer layer of the organoids. (E) Detection of marker proteins representing various cell types within the epithelial layer, including goblet cells (MUC3, MUC4), enterocytes (Villin), Paneth cells (DEFA5), enteroendocrine cells (CHGA), and colonic stem cells (LGR5). (F) Swelling assay confirming the presence of functional epithelial cells in the organoids treated after forskolin. All scale bars=50 μm. CO: colonic organoid.
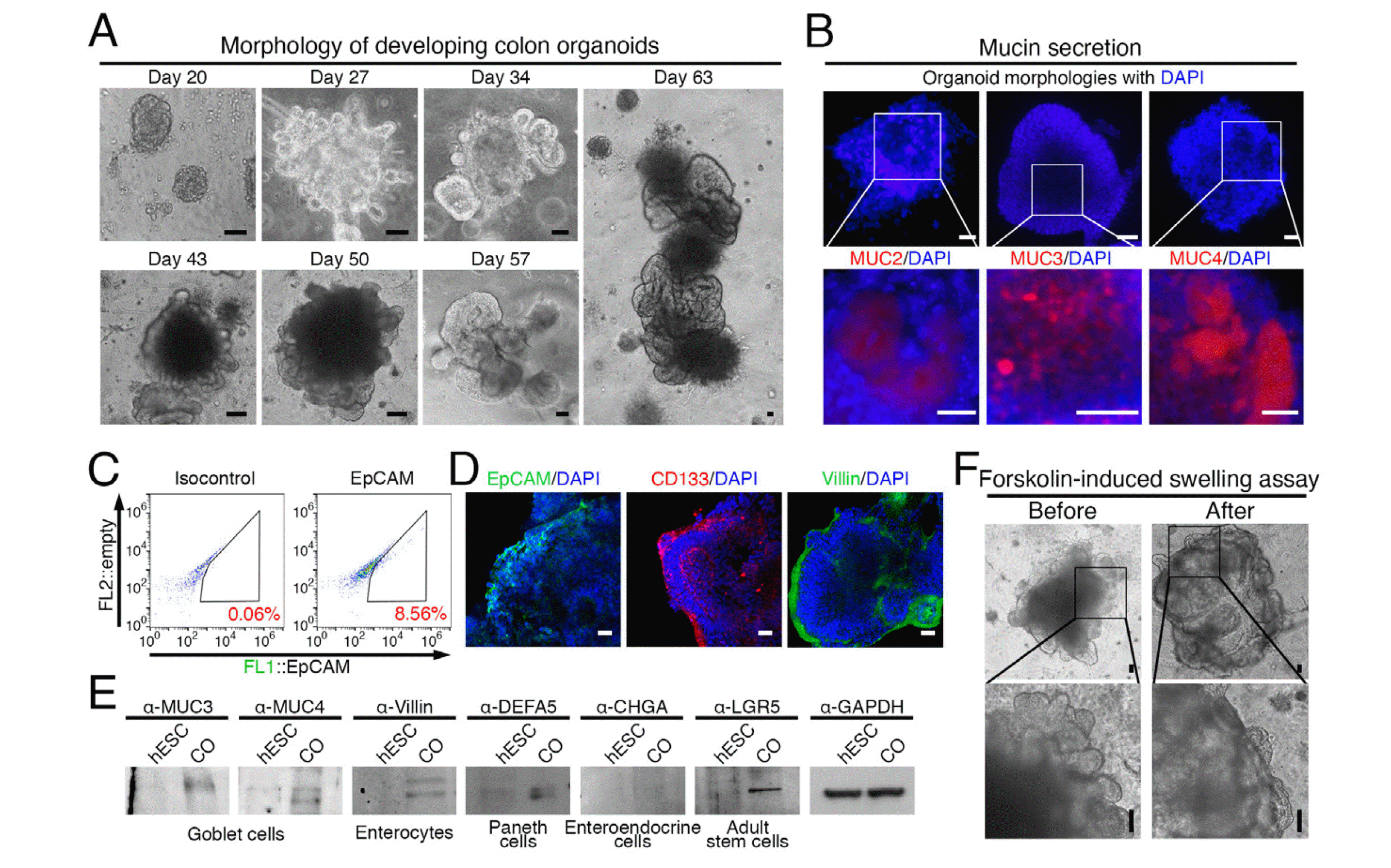
Fig. 3
Reciprocal interaction between colonic organoid and NCSC-derived peripheral neurons. (A) Schematic representation of co-culture of colon organoids with neural crest stem cells (NCSCs). (B) Representative images of NCSCs showing neuronal differentiation after 7 days of co-culture with colon organoids. (C) Representative FACS plot for NCSCs expressing PHOX2B after co-culture with colonic organoids. (D) Increased expression of PHOX2B and NDRG4 in FACS-isolated NCSCs after co-culture with colon organoids. (E) Enrichment of mRNA expression for LMX1A and CHGA in colon organoids after co-culture with NCSCs. (F) Schematic representation of transwell co-culture system. (G) Percentage of PHOX2B-expressing NCSCs induced by co-culture with colon organoids using direct contact or transwells. (H) Verification of PHOX2B-expressing cells after co-culture with colon organoids. (I) Expression of NDRG4 and LMX1A in colon organoids after co-culture with NCSCs using transwells. (J) Differentiated neuronal cells expressing TUJ1 containing serotonin (lower panel) and epithelial cells of colon organoids labeled with eGFP. Columnar morphology of epithelial cells are indicated by dotted red line in the right panel. (K) Proportion of EpCAM-expressing epithelial cells in colon organoids after co-culture with NCSCs. (L) Representative immunostaining image of Ki-67-expressing proliferating cells within colon organoids after co-culture with NCSCs, and the counting result. (M) Representative FACS plot for the isolation of EpCAM-expressing cells of colonic organoid, and representative images for attached cells after re-plating. (N) SOX9 expression in proliferating cells from EpCAM-expressing cells after 10 days of re-cultivation. Data expressed as mean±SD (n=9, 12 for G, biological repeat). p-values calculated by unpaired t-test (D, E, H, I, K, L) or one-way ANOVA (G). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant. All scale bars=50 μm. CO: colonic organoid.
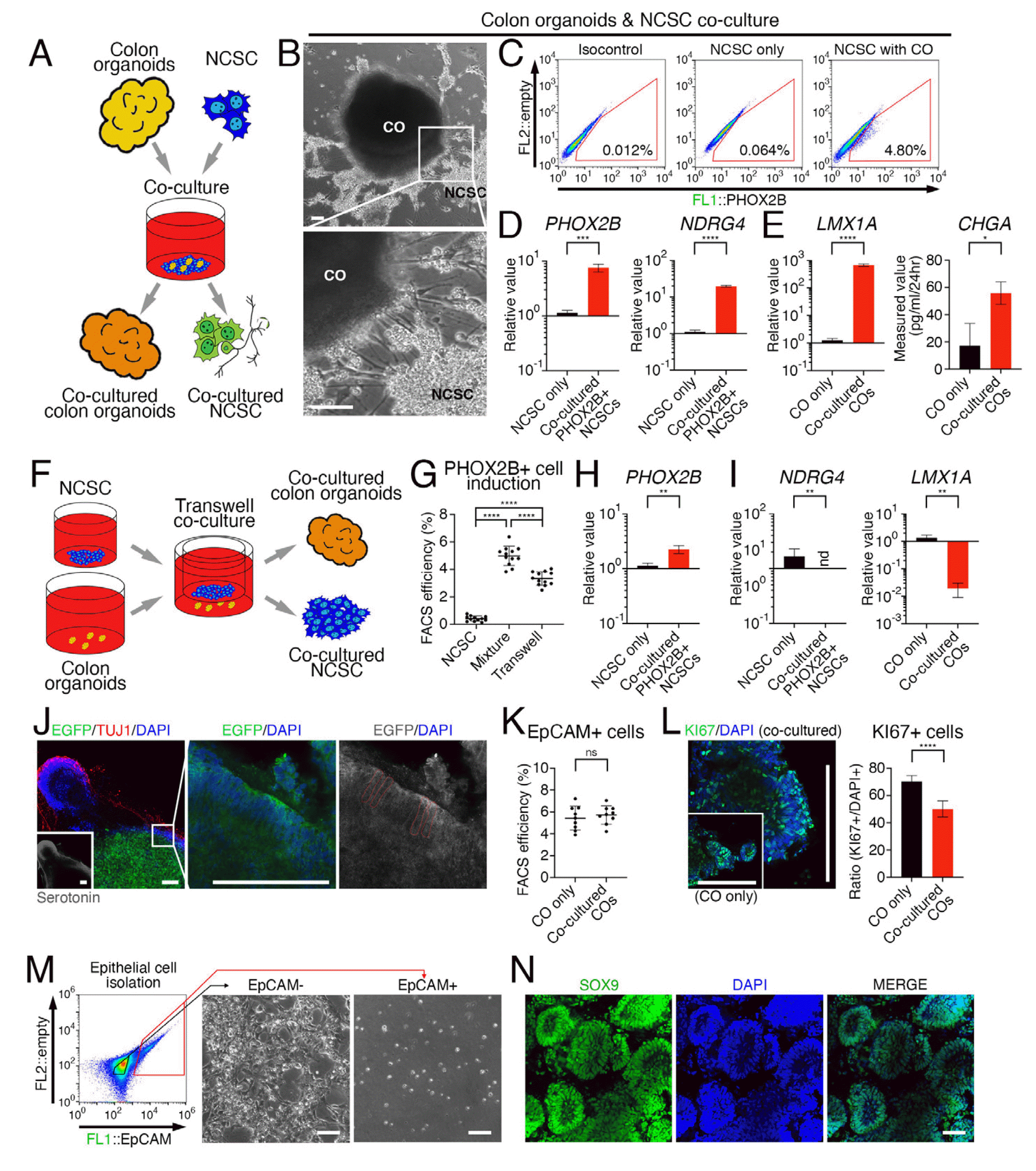
Fig. 4
Developing neurons influence colonic organoids. (A) Tracking the expression changes of PHOX2B and NDRG4 in NCSCs and LMX1A in colonic organoids according to the co-culture period of colonic organoids and NCSCs. (B) Immunostaining images of NCSC-derived neurons expressing TUJ1 as a key difference between co-culture time points of 4 and 6 days. (C) Significant increase in substance P in the co-culture medium on day 6 compared day 4, as measured by ELISA. (D) Expression of colonic epithelial cell marker proteins of colonic organoid after substance P administration were validated by immunoblotting. (E) Scattering distribution of cells expressing CHGA in the colonic organoid was confirmed using immunofluorescence staining. (F) Presence of enteroendocrine cells detectable by 5-HT within the colonic organoid, and columnar epithelial cell morphology (red dotted line). (G) The expression of NK1R and NK3R mRNA in colonic organoids was validated using qRT-PCR, with or without treatment with substance P. (H) Protein expression levels of CHGA in colonic organoids were measured using ELISA after inhibition of NK1R with specific antagonists. Data expressed as mean±SD (n=s, biological repeat). p-values calculated by two-way ANOVA (A and C) or one-way ANOVA (G and H). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant. Scale bars=25 μm (B and E) or 10 μm (F). CO: colonic organoid, SP: substance P.
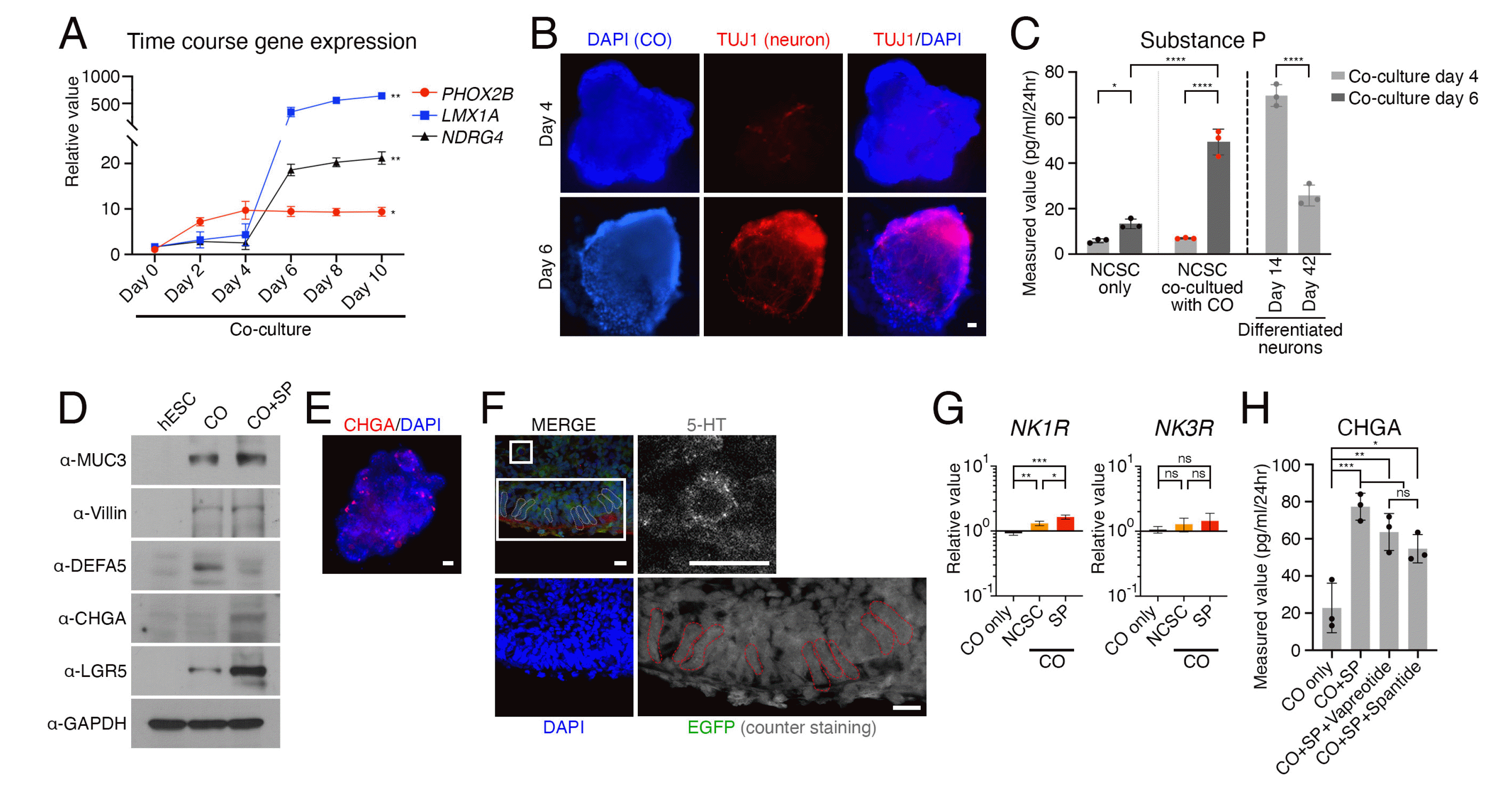
Table 1
List of primers and antibodies
| Inhihitors | ||
|---|---|---|
| Target | Company | Application |
| Vapreotide | MCE, HY-P3802 | NK1R antagonist |
| Spantide II | MCE, HY-P1722 | NK1R antagonist |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download