1. Tabak AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimaki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009; 373:2215–21.
2. Heianza Y, Arase Y, Fujihara K, Hsieh SD, Saito K, Tsuji H, et al. Longitudinal trajectories of HbA1c and fasting plasma glucose levels during the development of type 2 diabetes: the Toranomon Hospital Health Management Center Study 7 (TOPICS 7). Diabetes Care. 2012; 35:1050–2.
3. Meigs JB, Muller DC, Nathan DM, Blake DR, Andres R; Baltimore Longitudinal Study of Aging. The natural history of progression from normal glucose tolerance to type 2 diabetes in the Baltimore Longitudinal Study of Aging. Diabetes. 2003; 52:1475–84.
4. Chia CW, Egan JM, Ferrucci L. Age-related changes in glucose metabolism, hyperglycemia, and cardiovascular risk. Circ Res. 2018; 123:886–904.
5. Zhang X, Gregg EW, Williamson DF, Barker LE, Thomas W, Bullard KM, et al. A1C level and future risk of diabetes: a systematic review. Diabetes Care. 2010; 33:1665–73.
6. Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to singlevariant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018; 50:1505–13.
7. Flannick J, Mercader JM, Fuchsberger C, Udler MS, Mahajan A, Wessel J, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019; 570:71–6.
8. Liu CT, Raghavan S, Maruthur N, Kabagambe EK, Hong J, Ng MC, et al. Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet. 2016; 99:56–75.
9. Sikorska K, Rivadeneira F, Groenen PJ, Hofman A, Uitterlinden AG, Eilers PH, et al. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat Med. 2013; 32:165–80.
10. Little RJA, Rubin DB. Statistical analysis with missing data. 3rd ed. Hoboken: John Wiley & Sons;2020.
11. Ohn JH, Kwak SH, Cho YM, Lim S, Jang HC, Park KS, et al. 10-Year trajectory of β-cell function and insulin sensitivity in the development of type 2 diabetes: a community-based prospective cohort study. Lancet Diabetes Endocrinol. 2016; 4:27–34.
12. Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019; 9:1382.
13. Seo S, Park K, Lee JJ, Choi KY, Lee KH, Won S. SNP genotype calling and quality control for multi-batch-based studies. Genes Genomics. 2019; 41:927–39.
14. Song YE, Lee S, Park K, Elston RC, Yang HJ, Won S. ONETOOL for the analysis of family-based big data. Bioinformatics. 2018; 34:2851–3.
15. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164.
16. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009; 5:e1000529.
17. Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing;2013.
18. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient metaanalysis of genomewide association scans. Bioinformatics. 2010; 26:2190–1.
19. Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000; 66:1933–44.
20. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011; 88:76–82.
21. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013; 45:580–5.
22. Dominguez V, Raimondi C, Somanath S, Bugliani M, Loder MK, Edling CE, et al. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem. 2011; 286:4216–25.
23. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014; 46:310–5.
24. Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995; 310:170.
25. Gauderman WJ. Sample size requirements for matched casecontrol studies of gene-environment interaction. Stat Med. 2002; 21:35–50.
26. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010; 42:105–16.
27. Kim YJ, Go MJ, Hu C, Hong CB, Kim YK, Lee JY, et al. Largescale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011; 43:990–5.
28. Li X, Shu YH, Xiang AH, Trigo E, Kuusisto J, Hartiala J, et al. Additive effects of genetic variation in GCK and G6PC2 on insulin secretion and fasting glucose. Diabetes. 2009; 58:2946–53.
29. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009; 41:77–81.
30. Hu C, Zhang R, Wang C, Yu W, Lu J, Ma X, et al. Effects of GCK, GCKR, G6PC2 and MTNR1B variants on glucose metabolism and insulin secretion. PLoS One. 2010; 5:e11761.
31. Furlotte NA, Eskin E, Eyheramendy S. Genome-wide association mapping with longitudinal data. Genet Epidemiol. 2012; 36:463–71.
32. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018; 50:1412–25.
33. Gazal S, Loh PR, Finucane HK, Ganna A, Schoech A, Sunyaev S, et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat Genet. 2018; 50:1600–7.
34. Zhu Z, Wang X, Li X, Lin Y, Shen S, Liu CL, et al. Genetic overlap of chronic obstructive pulmonary disease and cardiovascular disease-related traits: a large-scale genome-wide cross-trait analysis. Respir Res. 2019; 20:64.
35. Pirruccello JP, Chaffin MD, Chou EL, Fleming SJ, Lin H, Nekoui M, et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat Genet. 2022; 54:40–51.
36. Johmura Y, Watanabe K, Kishimoto K, Ueda T, Shimada S, Osada S, et al. Fad24 causes hyperplasia in adipose tissue and improves glucose metabolism. Biol Pharm Bull. 2009; 32:1656–64.
37. Tominaga K, Johmura Y, Nishizuka M, Imagawa M. Fad24, a mammalian homolog of Noc3p, is a positive regulator in adipocyte differentiation. J Cell Sci. 2004; 117(Pt 25)(Pt 25):6217–26.
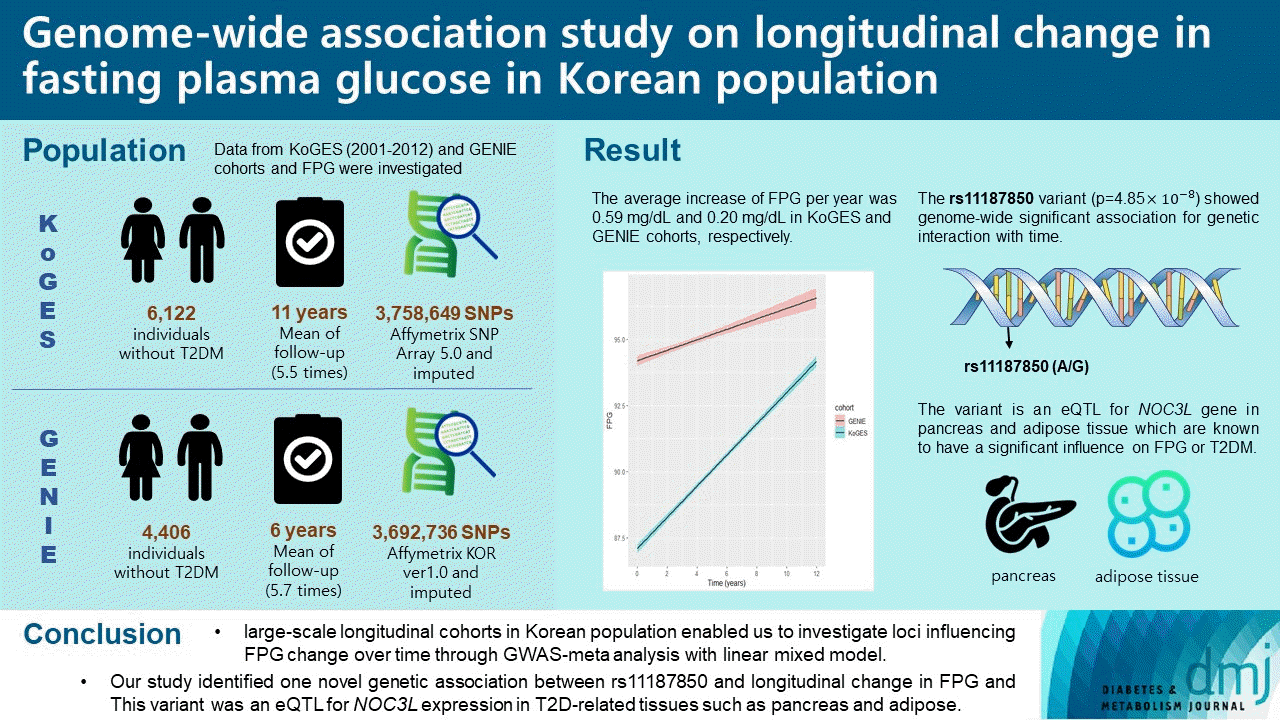
38. Liu CT, Merino J, Rybin D, DiCorpo D, Benke KS, BraggGresham JL, et al. Genome-wide association study of change in fasting glucose over time in 13,807 non-diabetic European ancestry individuals. Sci Rep. 2019; 9:9439.
39. Kwak SH, Chae J, Lee S, Choi S, Koo BK, Yoon JW, et al. Nonsynonymous variants in PAX4 and GLP1R are associated with type 2 diabetes in an East Asian population. Diabetes. 2018; 67:1892–902.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download