Abstract
Ion channels regulate a large number of cellular functions and their functional role in many diseases makes them potential therapeutic targets. Given their diverse distribution across multiple organs, the roles of ion channels, particularly in age-associated transcriptomic changes in specific organs, are yet to be fully revealed. Using RNA-seq data, we investigated the rat transcriptomic profiles of ion channel genes across 11 organs/tissues and 4 developmental stages in both sexes of Fischer 344 rats and identify tissue-specific and age-dependent changes in ion channel gene expression. Organ-enriched ion channel genes were identified. In particular, the brain showed higher tissue-specificity of ion channel genes, including Gabrd, Gabra6, Gabrg2, Grin2a, and Grin2b. Notably, age-dependent changes in ion channel gene expression were prominently observed in the thymus, including in Aqp1, Clcn4, Hvcn1, Itpr1, Kcng2, Kcnj11, Kcnn3, and Trpm2. Our comprehensive study of ion channel gene expression will serve as a primary resource for biological studies of aging-related diseases caused by abnormal ion channel functions.
The aging process is characterized by a cascade of cellular alterations and gradual deterioration in an organism’s metabolism. An increasing number of studies have highlighted the importance of exploring age-related diseases, including neurodegenerative and cardiovascular diseases [1,2]. Genome-wide gene expression analysis can provide insights into the molecular changes that underlie diseases, leading to a greater understanding of human biology [3]. Age-associated transcriptional alterations in cardiomyocytes isolated from 4- and 2-month old mice identified reduced activity of the mitochondrial electron transport system and cardiac-specific transcription regulators in older mice [4]. In addition, a recent study showed that transcriptional changes associated with aging were prominent in endothelial cells, and were directly related to functional changes in the aged brain [5].
Rats are larger in size, which exhibits advantages in the regard of handling and sampling, particularly in various surgical challenges. Indeed, genetic studies in rats have enabled genome-wide comparisons across species, and this could ultimately improve the transition of research to advance therapeutics for human diseases [6-8]. A RNA-seq BodyMap database showing organ-, age-, and sex-specific differential expression in 11 organs of rats ranging from juvenile to old age has been established [7,9]. Here, we use this valuable transcriptomic dataset [7,9] and our published computational pipeline, Sailfish-cir [10] to investigate the expression profile of ion channel genes across different organs/tissues including adrenal gland, brain, heart, kidney, liver, lung, muscle, spleen, thymus, testes, and uterus, and developmental stages (2-/6-/21-/104-week) in both sexes of Fischer 344 rats. Based on the knowledge of the diversity of ion channel subtypes and functions in entire tissues, we determined the extent to which ion channel genes showed a tissue-dependent pattern in the 11 organ/tissue types. The expression of several ion channel genes occurred in a tissue-specific manner; notably, the brain showed higher tissue-specific expression of ion channel genes than the other organs. We also found that the expression of ion channel genes in the thymus, including Aqp1, Clcn4, Hvcn1, Itpr1, Kcng2, Kcnj11, Kcnn3, and Trpm2, was positively correlated with age. We provide a comprehensive ion channel gene signature that occurs in tissue-specific and age-dependent manner.
The rat BodyMap RNA-seq data were downloaded from the NCBI Gene Expression Omnibus (GEO) database [11] with the accession number of GSE53960 [7,9]. The quality of the sequencing data was assessed by RNA-SeQC [12] and the raw sequencing reads were filtered by removing adaptor sequences, contamination, and low-quality reads. We quantified the expression levels for all the protein-coding genes according to the Ensembl rat gene annotation [13] (release 91) using the Sailfish-cir framework [14] with default settings.
Based on the rat BodyMap transcriptomic data, we computed the tissue specificity of each rat protein-coding gene. We applied the “tau” method developed by Yanai et al. [15] to calculate the tissue specificity index (TSI): , where TSI stands for the tissue specificity index; N represents the number of tissue types; and ei denotes the mean expression of a given gene in tissue i normalized to its maximum expression across all the tissues. A gene with high TSI implies this gene shows a strong tissue-specific expression.
All the statistical analyses were performed using the R statistical platform. Principal component analysis (PCA) was applied to visualize the tissue specificity of ion channel gene expression using the function “dudi.pca” in the R package “ade4”. The function “heatmap.2” in the R package “gplots” was employed to generate hierarchical clustering for the tissue-specific ion channel genes. The R generic functions, e.g., “t.test”, “ks.test”, “wilcox.test”, and “cor.test”, were applied to conduct t-test, Kolmogorov–Smirnov test, Wilcoxon test, and Spearman’s rank correlation test, respectively.
The definition of all the rat ion channel genes was obtained from the International Union of Basic and Clinical Pharmacology/British Pharmacological Society Guide to PHARMACOLOGY [16]. In total, 141 voltage-gated ion channel genes, 75 Ligand-gated ion channel genes, and 50 other ion channel genes were collected. To explore the dynamic expression pattern of ion channel genes, we obtained the rat RNA-seq transcriptomic BodyMap [7,9] from the GEO database [11] (GEO accession: GSE53960). Using our published RNA-seq quantification tool, Sailfish-cir [14], we summarized the ion channel gene expression profiles from the rat BodyMap dataset, which consists of 320 rat samples from 11 organ/tissue types, including adrenal gland, brain, heart, kidney, liver, lung, muscle, spleen, thymus, testes, and uterus across four developmental stages, i.e., 2-, 6-, 21- and 104-week. For each ion channel gene, the transcripts per million (TPM) was calculated to measure the expression level. We next investigated the top 30 highly expressed ion channel genes among the different tissue types. The ion channel genes that are among the top lists in at least eight tissue types were deemed “housekeeping”. In total, we identified 12 housekeeping ion channel genes, including Aqp1, Chrna7, Clcn3, Gja1, Itpr1, Itpr2, Mip, Orai1, P2rx4, Pkd2, Tpcn1, and Trpm7 (Fig. 1). The “housekeeping” ion channel genes defined by more/less stringent criteria were demonstrated in Supplementary Fig. 1 and 2, respectively.
We systematically investigated the tissue-specific expression for the ion channel genes. To understand the extent to which the expression of ion channel genes shows a tissue-dependent pattern, PCA upon ion channel gene expression was applied for all the samples. We found that the samples of the same organ/tissue type tended to aggregate together according to the first and second principal components (Fig. 2A), suggesting that ion channel genes are expressed in a strong tissue-specific manner. Similar to the previous observation [8], the brain samples showed an extremely unique PCA pattern compared with the other organ/tissue types (Fig. 2A), which was further confirmed by the fact that the mean TPM values of ion channel genes were significantly higher in the brain compared with the other tissue types (Kolmogorov–Smirnov test: p < 10–9; Fig. 2B). We further investigated the difference in tissue specificity between the ion channel genes and the other genes in rat transcriptome. We calculated TSI (see Methods for details) for both the ion channel genes and the other genes. We found that the TSI of ion channel genes was significantly higher than that of the other rat genes (Wilcoxon test: p < 10–10; Fig. 2C), which suggests that, compared with the other genes, ion channel genes are expressed in a more tissue-specific manner.
We further investigate whether the expression of the observed tissue-specific ion channel genes is relevant to the physiological function of the specific organ/tissue. A hierarchical clustering analysis was performed on the expression of all the rat ion channel genes across all the organ/tissue samples [8]. We observed that the samples from the same organ/tissue tended to be clustered into a single group except for thymus and testes. Also, each organ/tissue type presented several unique tissue-specific ion channel blocks (Fig. 3A). In addition, we found that the tissue types with similar physiological functions, such as thymus and spleen, tended to group together and share several common ion channel blocks, which suggests the common role of spleen and thymus in building the immune system (Fig. 3A). In contrast, the ion channel genes of brain samples showed a very distinct expression pattern compared with the other tissues (Fig. 3A), which largely mirrored the findings from Fig. 2A and B. To understand the functions of ion channels in different organs/tissues, we next looked into the top tissue-unique ion channel genes for each organ/tissue type. We found that the prioritized tissue-unique ion channels were related to the physiological function of each specific organ/tissue (Fig. 3B). For example, GABAA receptors and ionotropic glutamate receptors are dominant in brain, which are known to be critical for neural communication [17]; aquaporins are among the top kidney-specific ion channels, which facilitate renal water transportation through cell membrane [18]; Catsper channels are exclusively expressed in testes, which are actively involved in spermatogenesis [19] (Fig. 3B). Taken together, the above observations suggest that the tissue-specific ion channel genes play a critical role to perform their physiological functions in the individual organs/tissues.
We further investigated the alterations in ion channel gene expression in rat tissues at four different developmental stages (2-, 6-, 21-, and 104-week). PCA suggests that there is clearly a distinct pattern in ion channel gene expression across the four different developmental stages in all the tissue types (Fig. 4). To prioritize the ion channel genes that are longitudinally associated with age, Spearman’s rank correlation test was performed to measure the relationship between ion channel gene expression and rat age. The correlation coefficient (ρ) between expression and age and corresponding p-value were calculated for all the ion channel genes. In total, 23, 8, 19, 19, 8, 21, 22, 10, 74, 4, and 2 ion channel genes were found to be positively correlated with age, while 23, 42, 22, 26, 12, 33, 40, 13, 12, 19, and 12 ion channel genes were negatively correlated with age (Spearman’s rank correlation test: adjusted p < 0.05) in adrenal gland, brain, heart, kidney, liver, lung, muscle, spleen, thymus, testes, and uterus, respectively (Supplementary Table 1). More interestingly, thymus appeared to have more ion channel genes with large ρ values relative to the other tissues (t-test: p < 10–10; Fig. 5A), which suggests that the expression of most ion channel genes in thymus tends to monotonically increase with age (Kolmogorov–Smirnov test: p < 10-10; Fig. 5B). As an example, Fig. 5C demonstrated the top ion channel genes showing the strongest positive correlation between expression and age (ρ > 0.9) in thymus, including Aqp1, Clcn4, Hvcn1, Itpr1, Kcng2, Kcnj11, Kcnn3, and Trpm2, which potentially contribute to the development or age-related regression in thymus.
The role of ion channel impairment and/or mutation in the pathophysiology of various diseases has received increased attention. Changes in ionic flux maintain the resting potential, regulate intracellular calcium, and transmit electrical signals; therefore, altered ion channel gene expression causes cellular dysfunction, leading to various diseases [20,21]. The ion channel genes are ubiquitously expressed and selectively distributed in tissues, and their differential distribution contributes to their diverse cellular processes and physiological functions. For example, the expression of sodium/potassium/calcium exchanger 3 (NCKX3), copper-transporting ATPase 1 (ATP7A) and copper uptake protein 1 (CTR1) are the highest in the kidney, duodenum, and liver, respectively [22]. In addition, the CACNA1F gene encodes the Cav1.4 channel, which is preferentially expressed in the retina and is necessary for retinal neural transmission [23].
The age-related changes are complicated that multiple studies have reported altered cytokines, ROS, Ca2+ homeostasis, and inflammation play major role in pathologies of ion channels [24-28]. Indeed, altered splicing mRNA and under or over-expressed mRNA with aging have been reported and RNA instability, epigenetic changes, and telomere shortening, are accompanied by changes in regulation of gene expression [29-32]. Dynamic RNA modification in gene expression regulation would be associated with altered ion channel expression with aging. The age-related changes of ion channel expression or function have been identified in human and mouse [33,34]. For example, dysregulation of Ca2+ channels have been identified in neurodegenerative disease like an Alzheimer’s disease and lower expression of Ca2+-activated K+ channels have been seen with aging in human [35,36]. However, as far as we know, limited research has been done using transcriptomic approach to understand aging related ion channel expression in mouse or human.
To our knowledge, we are among the first to carry out the comprehensive assessment of the transcriptional profile of ion channel genes in different organs/tissues and developmental stages. We found a stronger tissue-specific expression pattern of ion channel genes. In particular, five genes: including Gabrd, Gabra6, Gabrg2, Grin2a, and Grin2b, were enriched in the brain. GABAA receptors are a class of ligand-gated Cl- channels that mediate inhibitory neurotransmitters in the central nervous system. The GABAA receptor is expressed exclusively in the cerebellum, but also expressed in testis and CD4+ T cells [37,38]. Among the GABAA receptor subtypes, we found that Gabra6, Gabrd, and Gabrg2 were highly specific to the brain. The disruption of GABA transmission has been associated with different neurodegenerative diseases. Mutations in Gabra6, Gabrd, and Gabrg2 have been identified in patients susceptible to febrile seizures and genetic epilepsy [39-41]. In addition, Gabrd is a potential prognostic marker in cancer, as it shows lower expression in isocitrate dehydrogenase (IDH) wild-type tumors than in IDH mutant diffuse low-grade glioma [42], and upregulated expression in hepatocellular carcinoma and adenocarcinoma [43,44]. Both Grin2A and Grin2B, which encode the NMDA receptor subunits, cause variable neurodevelopmental phenotypes. Grin2A variants are predominantly associated with epilepsy, while Grin2B variants are commonly found in patients with developmental disorders [45,46].
Abnormal changes in ionic gradients and calcium homeostasis across the membrane can underlie age-related deterioration of cellular function. The age-related dysfunction of ion channels is known to impact organ failure, including the heart and brain [47-49]. For example, increases in NaV1.5, Navβ1, and CaV1.2, and decreases in Kv1.5 and HCN1 were seen in the sino-atrial node of the heart in old rats, and resulted in age-dependent impairment of cardiac function [2]. Age is also the strongest risk factor for neurodegenerative pathogenesis, and was associated with downregulated Cav3.1 T-type calcium channel and impaired calcium homeostasis in Alzheimer’s disease [50,51]. In addition, transcriptomic alterations that impact neurovascular function and neurodegenerative conditions were identified in endothelial cells of aged mouse brain compared with young adult mouse brain [5].
One of the progressive, age-related declines the immune system is the regression of the thymus, which is represented by a decrease in size and impaired function [52]. The thymus is the primary lymphoid organ where T cells develop and recognize antigens. Disrupted thymic architecture, with reduced medullary size and impaired epithelial cells, was observed in the aged thymus [53,54]. In addition, microarray analyses performed on thymus from young, middle-aged, and old mice identified age-associated transcriptional changes [55]. Notably, in our transcriptomic profile of ion channel genes, age-dependent changes in expression of ion channel genes, including Aqp1, Clcn4, Hvcn1, Itpr1, Kcng2, Kcnj11, Kcnn3, and Trpm2 were observed in the thymus. Potassium channels, Kcng2, Kcnj11, and Kcnn3, allow K+ flux and are essential for regulating cellular excitability, and the critical role of these ion channels in disease has been highlighted. For example, the age-associated elevated expression of Kcnn3, small-conductance calcium-activated potassium (SK3) channels in the hippocampus of aged mice resulted in learning and memory deficits [56,57]. The Kcnj11 gene and Kir6.2 subunit of the adenosine triphosphate-sensitive potassium (KATP) channel is considered a promising candidate for identifying type2 diabetes risk because Kcnj11 is crucial for insulin secretion [58]. Aquaporin-1 (AQT1) channels in the major intrinsic protein family mediate water movement across the membrane and are expressed in thymocytes, where they play a critical role in decreasing apoptotic volume [59]. Decreased expression of Itpr1, which encodes the inositol-triphophate receptor type, was identified in aged human skeletal muscle [60]. Although an age-dependent change in ion channel gene expression was observed in the thymus, there is still limited knowledge about ion channel function associated with aged thymus.
In summary, we have developed a comprehensive platform for studying the rat transcriptome that provides tissue-specific and age-dependent ion channel gene expression information. The expression of several ion channels is highly tissue-specific, supporting the idea that a therapeutic target for a specific ion channel should take into account a particular organ. Further studies are required to investigate and validate these findings.
Supplementary data including one table and two figures can be found with this article online at https://doi.org/10.4196/kjpp.2023.27.1.85.
Notes
REFERENCES
1. Han K, Jin X, Guo X, Cao G, Tian S, Song Y, Zuo Y, Yu P, Gao G, Chang YZ. 2021; Nrf2 knockout altered brain iron deposition and mitigated age-related motor dysfunction in aging mice. Free Radic Biol Med. 162:592–602. DOI: 10.1016/j.freeradbiomed.2020.11.019. PMID: 33248265.
2. Tellez JO, Mczewski M, Yanni J, Sutyagin P, Mackiewicz U, Atkinson A, Inada S, Beresewicz A, Billeter R, Dobrzynski H, Boyett MR. 2011; Ageing-dependent remodelling of ion channel and Ca2+ clock genes underlying sino-atrial node pacemaking. Exp Physiol. 96:1163–1178. DOI: 10.1113/expphysiol.2011.057752. PMID: 21724736.
3. Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segrè AV, Djebali S, Niarchou A, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, et al. GTEx Consortium. 2015; Human genomics. The human transcriptome across tissues and individuals. Science. 348:660–665. DOI: 10.1126/science.aaa0355. PMID: 25954002. PMCID: PMC4547472. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84929015296&origin=inward.
4. Bodyak N, Kang PM, Hiromura M, Sulijoadikusumo I, Horikoshi N, Khrapko K, Usheva A. 2002; Gene expression profiling of the aging mouse cardiac myocytes. Nucleic Acids Res. 30:3788–3794. DOI: 10.1093/nar/gkf497. PMID: 12202764. PMCID: PMC137419. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0036713499&origin=inward.
5. Zhao L, Li Z, Vong JSL, Chen X, Lai HM, Yan LYC, Huang J, Sy SKH, Tian X, Huang Y, Chan HYE, So HC, Ng WL, Tang Y, Lin WJ, Mok VCT, Ko H. 2020; Pharmacologically reversible zonation-dependent endothelial cell transcriptomic changes with neurodegenerative disease associations in the aged brain. Nat Commun. 11:4413. DOI: 10.1038/s41467-020-18249-3. PMID: 32887883. PMCID: PMC7474063. PMID: 0cd6b88e4d0b4038b2b9807cac7e8608. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85090286333&origin=inward.
6. Showmaker KC, Cobb MB, Johnson AC, Yang W, Garrett MR. 2020; Whole genome sequencing and novel candidate genes for CAKUT and altered nephrogenesis in the HSRA rat. Physiol Genomics. 52:56–70. DOI: 10.1152/physiolgenomics.00112.2019. PMID: 31841396. PMCID: PMC6985787. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85078551518&origin=inward.
7. Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, et al. 2014; A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 5:3230. DOI: 10.1038/ncomms4230. PMID: 24510058. PMCID: PMC3926002. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84907492839&origin=inward.
8. Zhou T, Xie X, Li M, Shi J, Zhou JJ, Knox KS, Wang T, Chen Q, Gu W. 2018; Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA. 24:1443–1456. DOI: 10.1261/rna.067132.118. PMID: 30093490. PMCID: PMC6191709. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85055073493&origin=inward.
9. Yu Y, Zhao C, Su Z, Wang C, Fuscoe JC, Tong W, Shi L. 2014; Comprehensive RNA-Seq transcriptomic profiling across 11 organs, 4 ages, and 2 sexes of Fischer 344 rats. Sci Data. 1:140013. DOI: 10.1038/sdata.2014.13. PMID: 25977771. PMCID: PMC4381750. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84960885434&origin=inward.
10. Li M, Xie X, Zhou J, Sheng M, Yin X, Ko EA, Zhou T, Gu W. 2017; Quantifying circular RNA expression from RNA-Seq data using model-based framework. Bioinformatics. 33:2131–2139. DOI: 10.1093/bioinformatics/btx129. PMID: 28334396. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85020877053&origin=inward.
11. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. 2013; NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41:D991–D995. DOI: 10.1093/nar/gks1193. PMID: 23193258. PMCID: PMC3531084. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84874271270&origin=inward.
12. DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. 2012; RNA-SeQC: RNA-Seq metrics for quality control and process optimization. Bioinformatics. 28:1530–1532. DOI: 10.1093/bioinformatics/bts196. PMID: 22539670. PMCID: PMC3356847. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84861743958&origin=inward.
13. Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, et al. 2015; Ensembl 2015. Nucleic Acids Res. 43:D662–D669. DOI: 10.1093/nar/gku1010. PMID: 25352552. PMCID: PMC4383879. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84946037477&origin=inward.
14. Kim YW, Ko EA, Jung SC, Lee D, Seo Y, Kim S, Kim JH, Bang H, Zhou T, Ko JH. 2021; Transcriptomic insight into the translational value of two murine models in human atopic dermatitis. Sci Rep. 23:6616. DOI: 10.1038/s41598-021-86049-w. PMID: 33758305. PMCID: PMC7988112. PMID: 1c49ef5e932a4699b0cb9617720251bb. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85103202278&origin=inward.
15. Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O. 2005; Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 21:650–659. DOI: 10.1093/bioinformatics/bti042. PMID: 15388519. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=20144376345&origin=inward.
16. Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Southan C, Sharman JL, Campo B, Cavanagh DR, Alexander SPH, Davenport AP, Spedding M, Davies JA. 2020; The IUPHAR/BPS guide to PHARMACOLOGY in 2020: extending IMMUNOPHARMACOLOGY content and introducing the IUPHAR/MMV guide to Malaria PHARMACOLOGY. Nucleic Acids Res. 48:D1006–D1021. DOI: 10.1093/nar/gkz951. PMID: 31691834. PMCID: PMC7145572. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85077012757&origin=inward.
17. Petroff OA. 2002; GABA and glutamate in the human brain. Neuroscientist. 8:562–573. DOI: 10.1177/1073858402238515. PMID: 12467378. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0036892023&origin=inward.
18. Su W, Cao R, Zhang XY, Guan Y. 2020; Aquaporins in the kidney: physiology and pathophysiology. Am J Physiol Renal Physiol. 318:F193–F203. DOI: 10.1152/ajprenal.00304.2019. PMID: 31682170. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85077761313&origin=inward.
19. Sun XH, Zhu YY, Wang L, Liu HL, Ling Y, Li ZL, Sun LB. 2017; The Catsper channel and its roles in male fertility: a systematic review. Reprod Biol Endocrinol. 15:65. DOI: 10.1186/s12958-017-0281-2. PMID: 28810916. PMCID: PMC5558725. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85028062232&origin=inward.
20. Wang R, Gurguis CI, Gu W, Ko EA, Lim I, Bang H, Zhou T, Ko JH. 2015; Ion channel gene expression predicts survival in glioma patients. Sci Rep. 5:11593. DOI: 10.1038/srep11593. PMID: 26235283. PMCID: PMC4522676. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84938517742&origin=inward.
21. Hamilton S, Terentyev D. 2019; Altered intracellular calcium homeostasis and arrhythmogenesis in the aged heart. Int J Mol Sci. 20:2386. DOI: 10.3390/ijms20102386. PMID: 31091723. PMCID: PMC6566636. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85066853743&origin=inward.
22. Ahn C, Choi JS, Jeung EB. 2018; Organ-specific expression of the divalent ion channel proteins NCKX3, TRPV2, CTR1, ATP7A, IREG1 and HEPH in various canine organs. Mol Med Rep. 18:1773–1781. DOI: 10.3892/mmr.2018.9148. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85049576225&origin=inward.
23. Mansergh F, Orton NC, Vessey JP, Lalonde MR, Stell WK, Tremblay F, Barnes S, Rancourt DE, Bech-Hansen NT. 2005; Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet. 14:3035–3046. DOI: 10.1093/hmg/ddi336. PMID: 16155113. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=27544503765&origin=inward.
24. Hirata Y, Nomura K, Kato D, Tachibana Y, Niikura T, Uchiyama K, Hosooka T, Fukui T, Oe K, Kuroda R, Hara Y, Adachi T, Shibasaki K, Wake H, Ogawa W. 2022; A Piezo1/KLF15/IL-6 axis mediates immobilization-induced muscle atrophy. J Clin Invest. 132:1–13. DOI: 10.1172/JCI154611. PMID: 35290243. PMCID: PMC9159676. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85130190069&origin=inward.
25. Kiselyov KK, Ahuja M, Rybalchenko V, Patel S, Muallem S. 2012; The intracellular Ca²+ channels of membrane traffic. Channels (Austin). 6:344–351. DOI: 10.4161/chan.21723. PMID: 22907062. PMCID: PMC3508773. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84868140120&origin=inward.
26. Kaufmann U, Shaw PJ, Kozhaya L, Subramanian R, Gaida K, Unutmaz D, McBride HJ, Feske S. 2016; Selective ORAI1 inhibition ameliorates autoimmune central nervous system inflammation by suppressing effector but not regulatory T cell function. J Immunol. 196:573–585. DOI: 10.4049/jimmunol.1501406. PMID: 26673135. PMCID: PMC4707123. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84954118152&origin=inward.
27. Zöphel D, Hof C, Lis A. 2020; Altered Ca2+ homeostasis in immune cells during aging: role of ion channels. Int J Mol Sci. 22:110. DOI: 10.3390/ijms22010110. PMID: 33374304. PMCID: PMC7794837. PMID: 3064d0972f914f42bfef8ef8fdd9f824. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85098703184&origin=inward.
28. Guzik TJ, Touyz RM. 2017; Oxidative stress, inflammation, and vascular aging in hypertension. Hypertension. 70:660–667. DOI: 10.1161/HYPERTENSIONAHA.117.07802. PMID: 28784646. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85030998179&origin=inward.
29. Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, Yaghootkar H, Dutta A, Murray A, Frayling TM, Guralnik JM, Bandinelli S, Singleton A, Ferrucci L, Melzer D. 2011; Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 10:868–878. DOI: 10.1111/j.1474-9726.2011.00726.x. PMID: 21668623. PMCID: PMC3173580. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=80052803450&origin=inward.
30. Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK. 2006; Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet. 2:e115. DOI: 10.1371/journal.pgen.0020115. PMCID: PMC1513263. PMID: 16789832. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33746621756&origin=inward.
31. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013; The hallmarks of aging. Cell. 153:1194–1217. DOI: 10.1016/j.cell.2013.05.039. PMID: 23746838. PMCID: PMC3836174. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84878864199&origin=inward.
32. Viñuela A, Brown AA, Buil A, Tsai PC, Davies MN, Bell JT, Dermitzakis ET, Spector TD, Small KS. 2018; Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum Mol Genet. 27:732–741. DOI: 10.1093/hmg/ddx424. PMID: 29228364. PMCID: PMC5886097. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85041516302&origin=inward.
33. DiFranco M, Yu C, Quiñonez M, Vergara JL. 2013; Age-dependent chloride channel expression in skeletal muscle fibres of normal and HSALR myotonic mice. J Physiol. 591:1347–1371. DOI: 10.1113/jphysiol.2012.246546. PMID: 23247112. PMCID: PMC3607876. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84874428820&origin=inward.
34. Kakae M, Miyanohara J, Morishima M, Nagayasu K, Mori Y, Shirakawa H, Kaneko S. 2019; Pathophysiological role of TRPM2 in age-related cognitive impairment in mice. Neuroscience. 408:204–213. DOI: 10.1016/j.neuroscience.2019.04.012. PMID: 30999030. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85064867322&origin=inward.
35. Camandola S, Mattson MP. 2011; Aberrant subcellular neuronal calcium regulation in aging and Alzheimer's disease. Biochim Biophys Acta. 1813:965–973. DOI: 10.1016/j.bbamcr.2010.10.005. PMID: 20950656. PMCID: PMC3032815. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=79955665348&origin=inward.
36. Trombetta-Lima M, Krabbendam IE, Dolga AM. 2020; Calcium-activated potassium channels: implications for aging and age-related neurodegeneration. Int J Biochem Cell Biol. 123:105748. DOI: 10.1016/j.biocel.2020.105748. PMID: 32353429. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85084943207&origin=inward.
37. Geigerseder C, Doepner R, Thalhammer A, Frungieri MB, Gamel-Didelon K, Calandra RS, Köhn FM, Mayerhofer A. 2003; Evidence for a GABAergic system in rodent and human testis: local GABA production and GABA receptors. Neuroendocrinology. 77:314–323. DOI: 10.1159/000070897. PMID: 12806177. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0038010291&origin=inward.
38. Tian J, Lu Y, Zhang H, Chau CH, Dang HN, Kaufman DL. 2004; Gamma-aminobutyric acid inhibits T cell autoimmunity and the development of inflammatory responses in a mouse type 1 diabetes model. J Immunol. 173:5298–5304. DOI: 10.4049/jimmunol.173.8.5298. PMID: 15470076. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=6344285328&origin=inward.
39. Balan S, Sathyan S, Radha SK, Joseph V, Radhakrishnan K, Banerjee M. 2013; GABRG2, rs211037 is associated with epilepsy susceptibility, but not with antiepileptic drug resistance and febrile seizures. Pharmacogenet Genomics. 23:605–610. DOI: 10.1097/FPC.0000000000000000. PMID: 24061200. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84886601313&origin=inward.
40. Kumar P, Sharma D. 2020; Ameliorative effect of curcumin on altered expression of CACNA1A and GABRD in the pathogenesis of FeCl3-induced epilepsy. Mol Biol Rep. 47:5699–5710. DOI: 10.1007/s11033-020-05538-9. PMID: 32803504. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85089458405&origin=inward.
41. Haerian BS, Baum L, Kwan P, Cherny SS, Shin JG, Kim SE, Han BG, Tan HJ, Raymond AA, Tan CT, Mohamed Z. 2016; Contribution of GABRG2 polymorphisms to risk of epilepsy and febrile seizure: a multicenter cohort study and meta-analysis. Mol Neurobiol. 53:5457–5467. DOI: 10.1007/s12035-015-9457-y. PMID: 26452361. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84944705310&origin=inward.
42. Zhang H, Zhang L, Tang Y, Wang C, Chen Y, Shu J, Zhang K. 2019; Systemic screening identifies GABRD, a subunit gene of GABAA receptor as a prognostic marker in adult IDH wild-type diffuse low-grade glioma. Biomed Pharmacother. 118:109215. DOI: 10.1016/j.biopha.2019.109215. PMID: 31545245. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85070984585&origin=inward.
43. Sarathi A, Palaniappan A. 2019; Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma. BMC Cancer. 19:663. DOI: 10.1186/s12885-019-5838-3. PMID: 31277598. PMCID: PMC6612102. PMID: 6f78d393eebc4b259e146be696ab6389. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85069267765&origin=inward.
44. Wu M, Kim KY, Park WC, Ryu HS, Choi SC, Kim MS, et al. 2020; Enhanced expression of GABRD predicts poor prognosis in patients with colon adenocarcinoma. Transl Oncol. 13:100861. DOI: 10.1016/j.tranon.2020.100861. PMID: 32891902. PMCID: PMC7484591. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85090244339&origin=inward.
45. Myers SJ, Yuan H, Kang JQ, Tan FCK, Traynelis SF, Low CM. 2019; Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000Res. 8:F1000 Faculty Rev-1940. DOI: 10.12688/f1000research.18949.1. PMID: 31807283. PMCID: PMC6871362. PMID: 01db0138bf5048dbb3442018b9248bcd. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85076171438&origin=inward.
46. Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortüm F, Fritsch A, Pientka FK, Hellenbroich Y, Kalscheuer VM, Kohlhase J, Moog U, Rappold G, Rauch A, Ropers HH, von Spiczak S, Tönnies H, Villeneuve N, et al. 2010; Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 42:1021–1026. DOI: 10.1038/ng.677. PMID: 20890276. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=78049329316&origin=inward.
47. Comelli M, Meo M, Cervantes DO, Pizzo E, Plosker A, Mohler PJ, Hund TJ, Jacobson JT, Meste O, Rota M. 2020; Rhythm dynamics of the aging heart: an experimental study using conscious, restrained mice. Am J Physiol Heart Circ Physiol. 319:H893–H905. DOI: 10.1152/ajpheart.00379.2020. PMID: 32886003. PMCID: PMC7654658. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85092749808&origin=inward.
48. Benkert J, Hess S, Roy S, Beccano-Kelly D, Wiederspohn N, Duda J, Simons C, Patil K, Gaifullina A, Mannal N, Dragicevic E, Spaich D, Müller S, Nemeth J, Hollmann H, Deuter N, Mousba Y, Kubisch C, Poetschke C, Striessnig J, et al. 2019; Cav2.3 channels contribute to dopaminergic neuron loss in a model of Parkinson's disease. Nat Commun. 10:5094. DOI: 10.1038/s41467-019-12834-x. PMID: 31704946. PMCID: PMC6841684. PMID: 8271fb3221094175818e53f6966e1e51. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85074714711&origin=inward.
49. Signore S, Sorrentino A, Borghetti G, Cannata A, Meo M, Zhou Y, Kannappan R, Pasqualini F, O'Malley H, Sundman M, Tsigkas N, Zhang E, Arranto C, Mangiaracina C, Isobe K, Sena BF, Kim J, Goichberg P, Nahrendorf M, Isom LL, et al. 2015; Late Na+ current and protracted electrical recovery are critical determinants of the aging myopathy. Nat Commun. 6:8803. DOI: 10.1038/ncomms9803. PMID: 26541940. PMCID: PMC4638135. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84946599747&origin=inward.
50. Rice RA, Berchtold NC, Cotman CW, Green KN. 2014; Age-related downregulation of the CaV3.1 T-type calcium channel as a mediator of amyloid beta production. Neurobiol Aging. 35:1002–1011. DOI: 10.1016/j.neurobiolaging.2013.10.090. PMID: 24268883. PMCID: PMC3939046. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84893814237&origin=inward.
51. Ye H, Jalini S, Mylvaganam S, Carlen P. 2010; Activation of large-conductance Ca2+-activated K+ channels depresses basal synaptic transmission in the hippocampal CA1 area in APP (swe/ind) TgCRND8 mice. Neurobiol Aging. 31:591–604. DOI: 10.1016/j.neurobiolaging.2008.05.012. PMID: 18547679. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77049103601&origin=inward.
52. Lustig A, Carter A, Bertak D, Enika D, Vandanmagsar B, Wood W, Becker KG, Weeraratna AT, Taub DD. 2009; Transcriptome analysis of murine thymocytes reveals age-associated changes in thymic gene expression. Int J Med Sci. 6:51–64. DOI: 10.7150/ijms.6.51. PMID: 19214242. PMCID: PMC2640475. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=60049090034&origin=inward.
53. Aw D, Taylor-Brown F, Cooper K, Palmer DB. 2009; Phenotypical and morphological changes in the thymic microenvironment from ageing mice. Biogerontology. 10:311–322. DOI: 10.1007/s10522-008-9182-2. PMID: 18931936. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=67349171030&origin=inward.
54. Gui J, Zhu X, Dohkan J, Cheng L, Barnes PF, Su DM. 2007; The aged thymus shows normal recruitment of lymphohematopoietic progenitors but has defects in thymic epithelial cells. Int Immunol. 19:1201–1211. DOI: 10.1093/intimm/dxm095. PMID: 17804689. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34848922760&origin=inward.
55. Ki S, Park D, Selden HJ, Seita J, Chung H, Kim J, Iyer VR, Ehrlich LIR. 2014; Global transcriptional profiling reveals distinct functions of thymic stromal subsets and age-related changes during thymic involution. Cell Rep. 9:402–415. DOI: 10.1016/j.celrep.2014.08.070. PMID: 25284794. PMCID: PMC4194175. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84907963337&origin=inward.
56. Blank T, Nijholt I, Kye MJ, Radulovic J, Spiess J. 2003; Small-conductance, Ca2+-activated K+ channel SK3 generates age-related memory and LTP deficits. Nat Neurosci. 6:911–912. DOI: 10.1038/nn1101. PMID: 12883553. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0042861455&origin=inward.
57. Jacobsen JP, Redrobe JP, Hansen HH, Petersen S, Bond CT, Adelman JP, Mikkelsen JD, Mirza NR. 2009; Selective cognitive deficits and reduced hippocampal brain-derived neurotrophic factor mRNA expression in small-conductance calcium-activated K+ channel deficient mice. Neuroscience. 163:73–81. DOI: 10.1016/j.neuroscience.2009.05.062. PMID: 19482064. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=68349144706&origin=inward.
58. Khan V, Verma AK, Bhatt D, Khan S, Hasan R, Goyal Y, Ramachandran S, Alsahli MA, Rahmani AH, Almatroudi A, Shareef MY, Meena B, Dev K. 2020; Association of genetic variants of KCNJ11 and KCNQ1 genes with risk of type 2 diabetes mellitus (T2DM) in the Indian population: a case-control study. Int J Endocrinol. 2020:5924756. DOI: 10.1155/2020/5924756. PMID: 33101408. PMCID: PMC7569458. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85094644340&origin=inward.
59. Jablonski EM, Webb AN, McConnell NA, Riley MC, Hughes FM Jr. 2004; Plasma membrane aquaporin activity can affect the rate of apoptosis but is inhibited after apoptotic volume decrease. Am J Physiol Cell Physiol. 286:C975–C985. DOI: 10.1152/ajpcell.00180.2003. PMID: 14644770. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=2142703737&origin=inward.
60. Choi JY, Hwang CY, Lee B, Lee SM, Bahn YJ, Lee KP, Kang M, Kim YS, Woo SH, Lim JY, Kim E, Kwon KS. 2016; Age-associated repression of type 1 inositol 1, 4, 5-triphosphate receptor impairs muscle regeneration. Aging (Albany NY). 8:2062–2080. DOI: 10.18632/aging.101039. PMID: 27658230. PMCID: PMC5076452. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84991494501&origin=inward.
Fig. 1
The “housekeeping” ion channel genes.
For each tissue type, the top 30 ion channel genes with the highest expression were prioritized. The genes highlighted in red are the “housekeeping” ion channel genes, which are among the top 30 list in at least eight tissue types, including Aqp1, Chrna7, Clcn3, Gja1, Itpr1, Itpr2, Mip, Orai1, P2rx4, Pkd2, Tpcn1, and Trpm7.
Fig. 2
Tissue specificity of ion channel gene expression.
(A) PCA of ion channel gene expression. Each dot represents one sample. PC1 stands the first principal component while PC2 is the second principal component. (B) Cumulative distribution of the mean TPM for different organ/tissue types. (C) Histogram of the TSI of the ion channel genes and the other genes. PCA, principal component analysis; TPM, transcripts per million; TSI, tissue specificity index.
Fig. 3
Map of tissue-specific ion channel genes.
(A) Hierarchical clustering of the ion channel genes. Each green point stands one tissue-specific ion channel gene. Red color represents relatively increased ion channel gene expression whereas blue color represents relatively lower expression. (B) The top tissue-unique ion channel genes. For each tissue type, the top five ion channel genes with the largest fold change in expression relative to the other tissues are listed. Darker red indicates a larger fold change, while lighter red indicates a smaller fold change. Gray indicates non-significant upregulation.
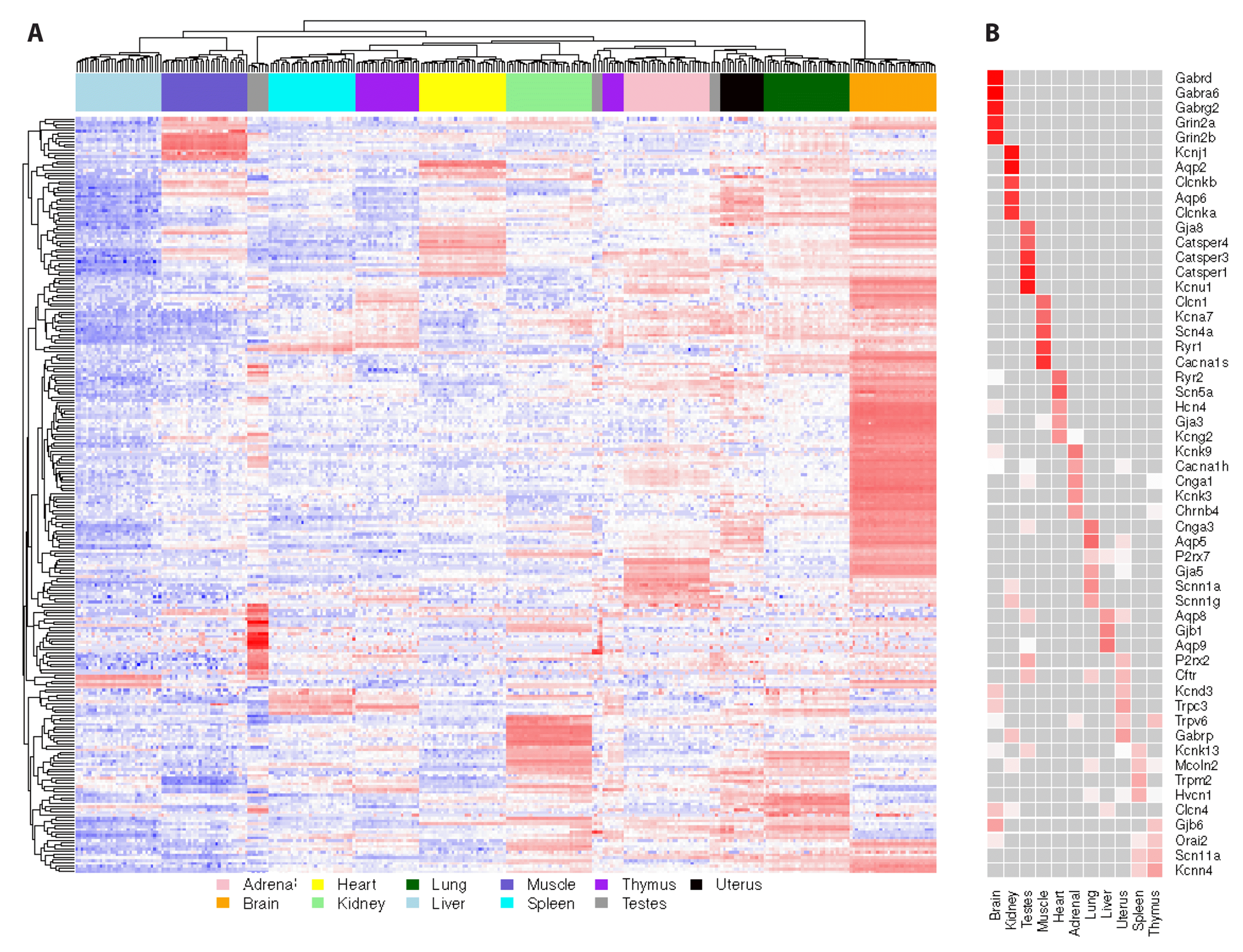
Fig. 4
Developmental stage-dependent ion channel gene expression.
PCA indicates that the expression of the ion channel genes shows a strong developmental stage-dependent manner. Each dot represents one sample in a given organ/tissue. PC1 stands the first principal component while PC2 is the second principal component. PCA, principal component analysis.
Fig. 5
Developmental stage-dependent ion channel genes in thymus.
(A) Distribution of the correlation coefficients (ρ) between the ion channel gene expression and age across the 11 organs/tissues. The ρ values were computed using Spearman’s rank correlation test. (B) Cumulative distribution of the ρ values across the 11 tissue types. There are more ion channel genes in thymus with strong positive correlation between expression and age compared with the other organs/tissues. (C) The top eight ion channel genes showing the strongest positive correlation between expression and age (ρ > 0.9) in thymus. TPM, transcripts per million.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download