Abstract
Background
Methods
Results
Conclusions
ACKNOWLEDGEMENTS
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Lee B, Hwang N, Park HD, and Lee SY. Data curation: Lee B, Heo WY, Lee HS, and Hwang N. Formal analysis: Lee B, Heo WY, Kim JA, Lee HS, and Lee SY. Investigation: Lee B, Heo WY, Lee HS, Hwang N, and Lee SY. Methodology: Lee B, Heo WY, Hwang N, Park HD, and Lee SY. Project administration: Lee SY. Resources: Hwang N, Park HD, Sung SI, Chang YS, Park WS, and Lee SY. Supervision: Park HD, Sung SI, Chang YS, Park WS, and Lee SY. Validation: Lee B, Heo WY, Kim JA, Lee HS, and Lee SY. Visualization: Lee B. Writing — original draft: Lee B. Writing — review and editing: Lee B and Lee SY. All authors accept responsibility for the entire content of this manuscript and approve its submission.
REFERENCES
Fig. 1
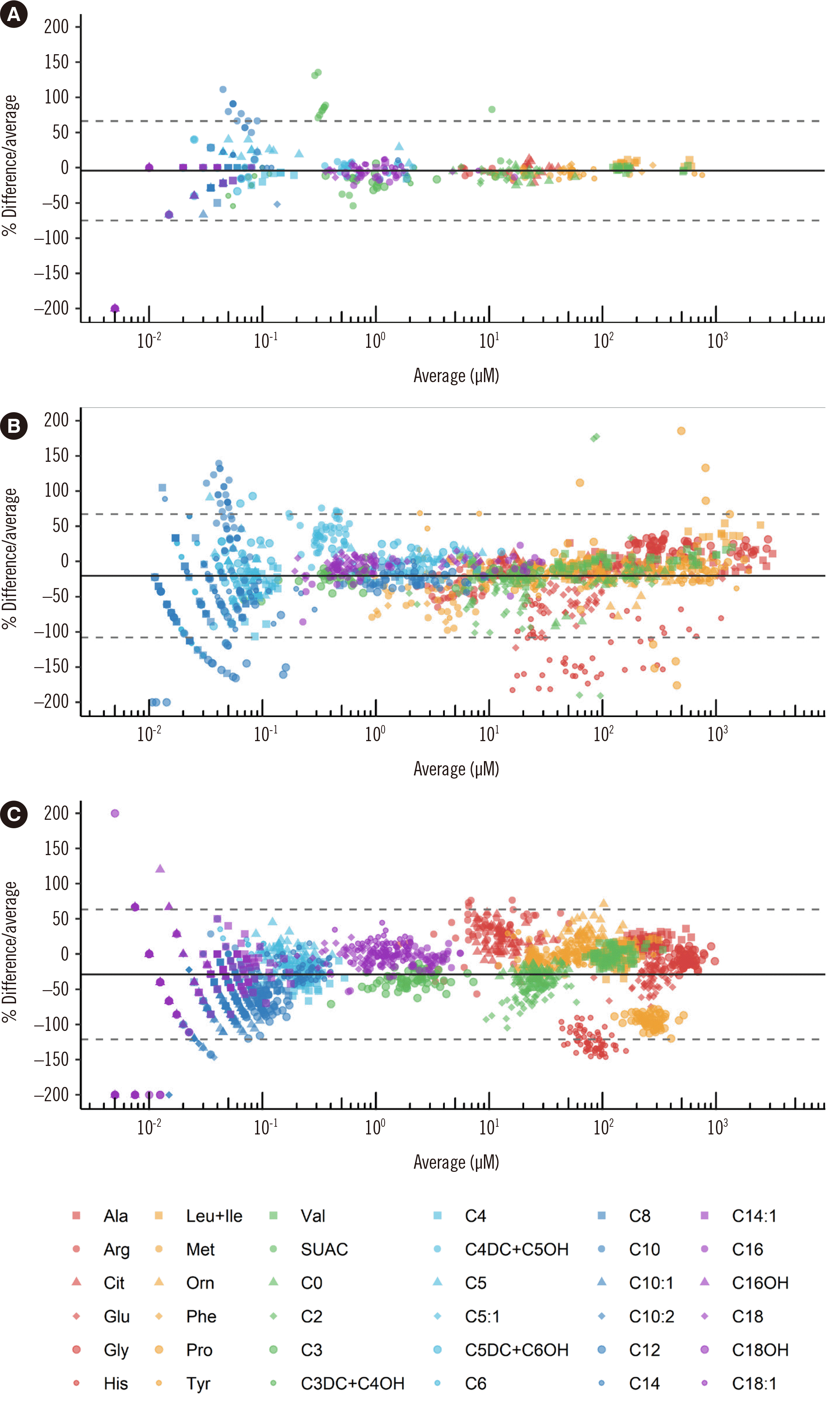
Fig. 2
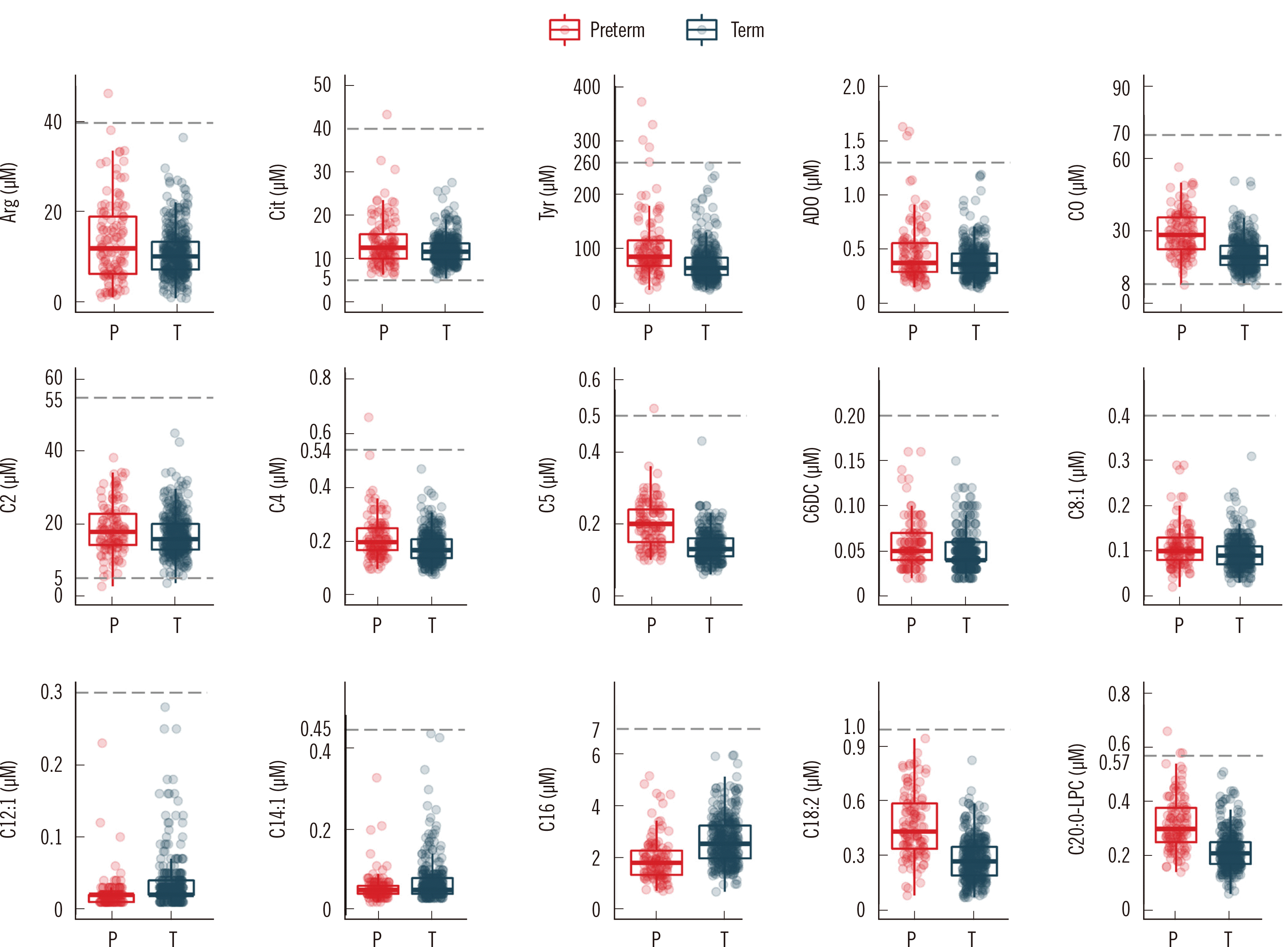
Fig. 3
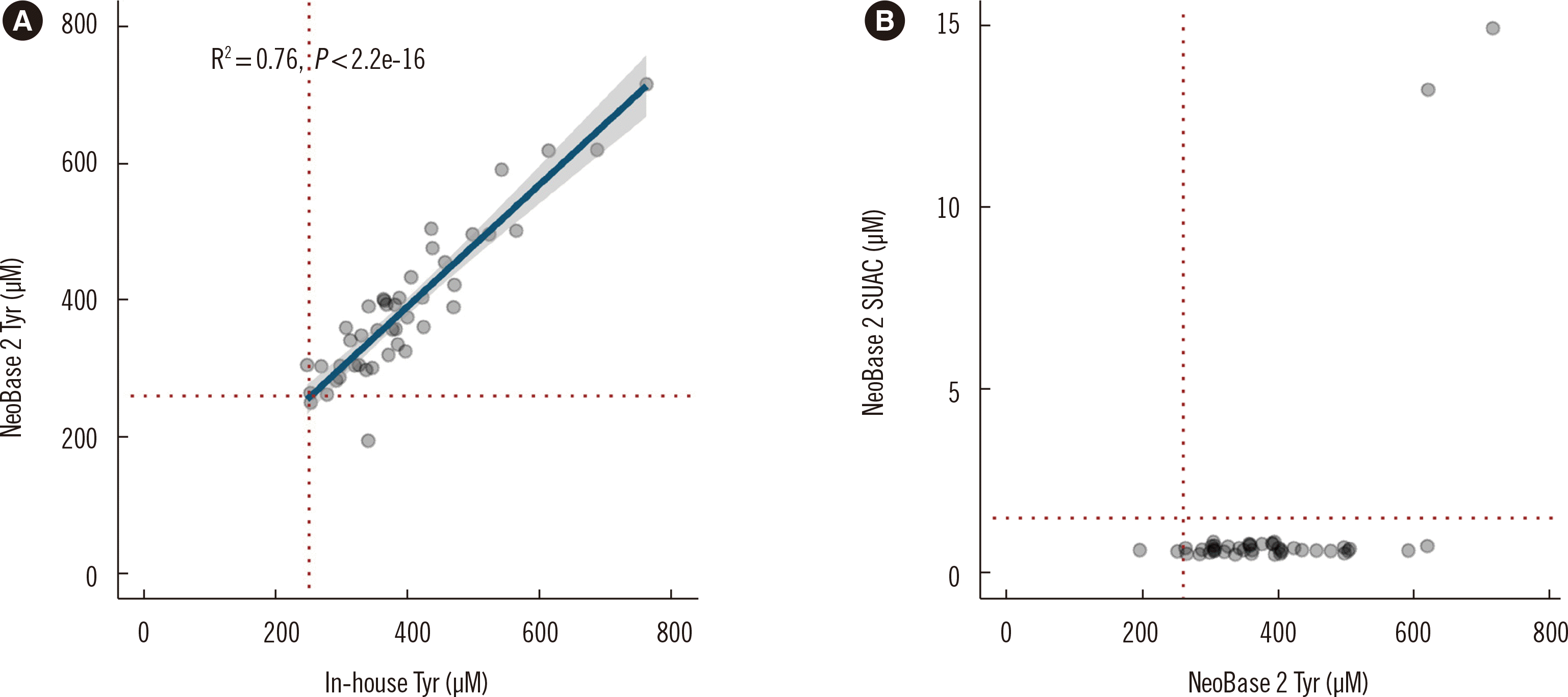
Table 1
| Analyte | LOD (μM) | LLOQ (μM) | Precision* (%CV) | Linearity | Recovery* (%) | Carryover (μM) | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Within-run | Between-day | Range (μM)† | R2 | |||||
| Ala | 1.13 | 2.26 | 5.06 ± 1.13 | 7.44 ± 2.90 | 398.40–2,710.77 | 0.9976 | 80.2 ± 13.2 | 3.45 |
| Arg | 0.17 | 0.33 | 3.46 ± 0.94 | 1.72 ± 1.59 | 6.03–1,311.86 | 0.9997 | 90.7 ± 3.8 | 0.65 |
| Cit | 0.21 | 0.42 | 5.10 ± 0.91 | 1.80 ± 2.06 | 23.44–917.48 | 0.9978 | 92.5 ± 7.4 | 0.23 |
| Gln + Lys | 0.46 | 0.92 | 4.50 ± 1.39 | 1.99 ± 1.68 | 2.57 | |||
| Glu | 0.17 | 0.33 | 4.56 ± 1.11 | 4.79 ± 1.99 | 0.14 | |||
| Gly | 2.16 | 4.31 | 4.27 ± 1.46 | 2.24 ± 1.86 | 424.46–2,791.61 | 0.9990 | 96.2 ± 13.6 | 8.42 |
| His | 0.18 | 0.35 | 5.16 ± 1.14 | 7.34 ± 1.80 | 0.63 | |||
| Leu + Ile | 0.21 | 0.43 | 4.14 ± 1.06 | 2.66 ± 1.60 | 242.94–1,224.43 | 0.9962 | 88.4 ± 16.1 | 1.74 |
| Met | 0.50 | 1.00 | 4.40 ± 1.27 | 5.56 ± 1.94 | 15.67–466.28 | 0.9981 | 81.5 ± 6.2 | 0.53 |
| Orn | 0.66 | 1.33 | 3.72 ± 1.05 | 2.52 ± 1.64 | 114.72–1,317.90 | 0.9984 | 79.1 ± 6.9 | 9.90 |
| Phe | 5.30 | 10.59 | 4.14 ± 1.21 | 3.43 ± 2.32 | 74.78–1,000.80 | 0.9986 | 83.9 ± 8.4 | 0.23 |
| Pro | 0.20 | 0.39 | 4.20 ± 1.20 | 5.56 ± 2.08 | 212.29–1,408.03 | 0.9976 | 74.6 ± 10.6 | 1.96 |
| Tyr | 0.52 | 1.05 | 3.66 ± 1.39 | 3.21 ± 2.10 | 78.77–1,452.52 | 0.9990 | 79.6 ± 7.7 | 0.62 |
| Val | 0.15 | 0.30 | 4.01 ± 0.85 | 4.00 ± 1.73 | 227.75–1,188.23 | 0.9960 | 87.2 ± 16.5 | 1.36 |
| ASA | 10.96 ± 2.08 | 13.90 ± 11.28 | 0.64 | |||||
| SUAC | 0.03 | 0.07 | 5.68 ± 1.98 | 3.62 ± 2.40 | 0.53–38.69 | 0.9995 | 61.0 ± 3.9 | 0.13 |
| ADO | 0.02 | 0.03 | 3.90 ± 0.14 | 4.13 ± 1.83 | < 0.01 | |||
| D-ADO | < 0.01 | |||||||
| C0 | 0.03 | 0.07 | 4.52 ± 1.14 | 3.37 ± 2.32 | 27.62–1,057.47 | 0.9995 | 83.6 ± 5.2 | 0.12 |
| C2 | 0.03 | 0.07 | 4.40 ± 0.87 | 1.93 ± 1.52 | 9.51–333.59 | 0.9984 | 94.0 ± 6.9 | 0.07 |
| C3 | 0.03 | 0.07 | 4.44 ± 0.85 | 1.85 ± 1.45 | 1.53–41.22 | 0.9990 | 93.3 ± 6.2 | 0.01 |
| C3DC + C4OH | 4.10 ± 0.71 | 4.29 ± 1.26 | < 0.01 | |||||
| C4 | 4.96 ± 0.76 | 1.30 ± 1.45 | 0.26–19.79 | 0.9991 | 87.0 ± 4.6 | 0.01 | ||
| C4DC + C5OH | 4.29 ± 1.35 | 2.48 ± 2.12 | 0.01 | |||||
| C5 | 4.71 ± 1.02 | 4.01 ± 2.25 | 0.14–18.05 | 0.9992 | 101.1 ± 4.8 | 0.02 | ||
| C5:1 | 5.97 ± 1.02 | 3.04 ± 2.54 | < 0.01 | |||||
| C5DC + C6OH | 5.81 ± 1.62 | 4.22 ± 1.18 | 0.06–39.85 | 0.9998 | 83.3 ± 2.4 | < 0.01 | ||
| C6 | 4.63 ± 1.04 | 2.15 ± 2.15 | 0.05–21.63 | 0.9994 | 88.5 ± 5.4 | < 0.01 | ||
| C6DC | < 0.01 | |||||||
| C8 | 4.85 ± 1.07 | 2.24 ± 2.08 | 0.04–17.51 | 0.9990 | 97.5 ± 6.3 | < 0.01 | ||
| C8:1 | < 0.01 | |||||||
| C10 | 5.71 ± 1.21 | 2.72 ± 0.81 | 0.03–17.04 | 0.9990 | 83.5 ± 5.1 | 0.04 | ||
| C10:1 | < 0.01 | |||||||
| C10:2 | < 0.01 | |||||||
| C12 | 5.47 ± 1.07 | 3.21 ± 2.17 | 0.03–21.44 | 0.9986 | 96.0 ± 6.2 | < 0.01 | ||
| C12:1 | < 0.01 | |||||||
| C14 | 5.15 ± 0.77 | 2.47 ± 2.29 | 0.11–20.17 | 0.9987 | 96.5 ± 6.4 | < 0.01 | ||
| C14OH | < 0.01 | |||||||
| C14:1 | 0.02 | 0.03 | 5.32 ± 0.97 | 2.20 ± 2.10 | < 0.01 | |||
| C14:2 | 0.02 | 0.03 | < 0.01 | |||||
| C16 | 0.02 | 0.03 | 5.05 ± 1.75 | 3.00 ± 2.21 | 1.13–54.88 | 0.9984 | 91.4 ± 7.2 | 0.01 |
| C16OH | 6.14 ± 2.82 | 4.43 ± 1.51 | < 0.01 | |||||
| C16:1 | < 0.01 | |||||||
| C16:1OH | 0.01 | |||||||
| C18 | 5.10 ± 1.81 | 2.75 ± 2.40 | 0.80–37.70 | 0.9981 | 92.5 ± 7.5 | 0.01 | ||
| C18OH | 7.23 ± 2.09 | 2.16 ± 2.73 | < 0.01 | |||||
| C18:1 | 5.93 ± 2.88 | 3.13 ± 3.21 | < 0.01 | |||||
| C18:1OH | < 0.01 | |||||||
| C18:2 | 6.54 ± 3.03 | 3.20 ± 3.11 | < 0.01 | |||||
| C18:2OH | 0.04 | |||||||
| C20 | < 0.01 | |||||||
| C22 | 0.01 | |||||||
| C24 | < 0.01 | |||||||
| C26 | 4.82 ± 1.57 | 2.19 ± 3.09 | 0.01 | |||||
| C20:0-LPC | 6.74 ± 2.87 | 4.13 ± 2.89 | < 0.01 | |||||
| C22:0-LPC | 6.41 ± 1.69 | 3.08 ± 2.92 | < 0.01 | |||||
| C24:0-LPC | 6.65 ± 1.88 | 1.59 ± 2.33 | < 0.01 | |||||
| C26:0-LPC | 9.07 ± 6.80 | 3.38 ± 2.98 | 0.02 | |||||
*The results of precision and recovery analyses are presented as mean±SD; †The mean values of the duplicate runs were used.
Abbreviations: LOD, limit of detection; LLOQ, lower limit of quantification; ASA, argininosuccinic acid; SUAC, succinylacetone; ADO, adenosine; D-ADO, 2´-deoxyadenosine; LPC, lysophosphatidylcholine; amino acids are abbreviated as three-letter symbols according to the International Union of Pure and Applied Chemistry (IUPAC) nomenclature.
Table 2
| Analyte | Accuracy (% agreement [total agreement rate, abnormality agreement rate]) | ||
|---|---|---|---|
|
|
|||
| NSQAP PT* | KEQAS PT* | In-house derivatized assay | |
| Ala | 100% (52/52, 14/14) | 100% (64/64, 0/0) | |
| Arg | 100% (20/20, 2/2) | 100% (54/54, 11/11) | 95.3% (61/64, 1/1) |
| Cit | 100% (20/20, 3/3) | 100% (54/54, 13/13) | 100% (64/64, 1/1) |
| Glu | 100% (49/49, 6/6) | 100% (64/64, 0/0) | |
| Gly | 98.1% (52/53, 12/12) | 100% (64/64, 2/2) | |
| His | 100% (49/49, 11/11) | 98.4% (63/64, 1/1) | |
| Leu + Ile | 100% (19/19, 4/4) | 100% (53/53, 20/20) | 100% (64/64, 3/3) |
| Met | 100% (20/20, 5/5) | 100% (54/54, 21/21) | 98.4% (63/64, 1/2) |
| Orn | 100% (51/51, 15/15) | 100% (64/64, 2/2) | |
| Phe | 100% (20/20, 3/3) | 100% (54/54, 21/21) | 100% (64/64, 1/1) |
| Pro | 100% (54/54, 16/17) | 100% (64/64, 0/0) | |
| Tyr | 100% (20/20, 2/2) | 100% (53/53, 19/19) | 100% (64/64, 1/1) |
| Val | 100% (20/20, 5/5) | 100% (52/52, 15/15) | 100% (64/64, 2/2) |
| SUAC | 100% (20/20, 2/2) | ||
| C0† | 100% (20/20, 1/1) | 100% (49/49, 18/18) | 100% (64/64, 1/1) |
| C2† | 100% (18/18, 0/0) | 100% (54/54, 18/18) | 98.4% (63/64, 0/1) |
| C3 | 100% (19/19, 1/1) | 100% (53/53, 16/16) | 100% (64/64, 2/2) |
| C3DC + C4OH | 100% (20/20, 3/3) | ||
| C4 | 100% (20/20, 1/1) | 100% (53/53, 12/12) | 100% (64/64, 0/0) |
| C4DC + C5OH | 100% (20/20, 2/2) | 98.1% (53/54, 6/6) | |
| C5 | 100% (20/20, 2/2) | 98.1% (53/54, 16/16) | 100% (64/64, 0/0) |
| C5:1 | 100% (20/20, 1/1) | 100% (64/64, 0/0) | |
| C5DC + C6OH | 100% (19/19, 1/1) | 100% (53/53, 17/17) | |
| C6 | 100% (19/19, 1/1) | 100% (54/54, 13/13) | 100% (64/64, 0/0) |
| C8 | 100% (20/20, 1/1) | 100% (54/54, 13/13) | 100% (64/64, 0/0) |
| C10 | 100% (20/20, 1/1) | 100% (54/54, 12/12) | 100% (64/64, 0/0) |
| C10:1 | 100% (20/20, 1/1) | 100% (64/64, 0/0) | |
| C10:2 | 100% (20/20, 1/1) | 100% (64/64, 0/0) | |
| C12 | 100% (54/54, 13/13) | 100% (64/64, 0/0) | |
| C14 | 100% (20/20, 4/4) | 100% (53/53, 13/13) | 100% (64/64, 0/0) |
| C14:1 | 100% (19/19, 3/3) | 100% (64/64, 0/0) | |
| C16 | 100% (20/20, 1/1) | 100% (54/54, 14/14) | 100% (64/64, 0/0) |
| C16OH | 100% (20/20, 2/2) | 100% (64/64, 0/0) | |
| C18 | 100% (20/20, 1/1) | 100% (53/53, 13/13) | 98.4% (63/64, 0/0) |
| C18OH | 100% (20/20, 2/2) | 100% (64/64, 0/0) | |
| C18:1 | 100% (20/20, 1/1) | 100% (64/64, 0/0) | |
*Blank results indicate that the corresponding analytes are not included in the PT program; †The lower cutoff, upper cutoff, and both lower and upper cutoffs were applied for comparison with the NSQAP PT, KEQAS PT, and in-house derivatized assay, respectively.
Abbreviations: ASA, argininosuccinic acid; SUAC, succinylacetone; ADO, adenosine; D-ADO, 2´-deoxyadenosine; LPC, lysophosphatidylcholine; KEQAS, Korean Association of External Quality Assessment Service; NSQAP, Newborn Screening Quality Assurance Program; PT, proficiency testing; amino acids are abbreviated as three-letter symbols according to the International Union of Pure and Applied Chemistry (IUPAC) nomenclature.
Table 3
| Analyte (μM) | Cutoff | Distribution (99.5 percentile [mean±SD]) | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Our lab | CLIR 50 percentile | NSQAP [20] | KEQAS [21] | Cho, et al. [22] | Preterm | Term | |
| Ala | 700 | 698.20 | 694 | 532.30 (261.79 ± 74.86) | 603.96 (266.15 ± 75.50) | ||
| Arg | 40 | 50 | 57.60 | 57.10 | 40 | 41.76 (13.84 ± 8.98) | 28.33 (11.24 ± 5.30) |
| Cit | 5 (L), 40 | 5 (L), 50 | 49.80 | 48.00 | 40 | 6.45*, 37.06 (13.67 ± 5.70) | 6.24*, 25.61 (11.95 ± 3.19) |
| Gln + Lys | 1,200 | 850 | 1,179.91 (600.04 ± 194.87) | 1,164.96 (654.53 ± 148.29) | |||
| Glu | 500 | 850 | 409.30 (239.99 ± 61.36) | 432.26 (224.09 ± 55.85) | |||
| Gly | 900 | 851 | 893.00 | 1,000 | 730.90 (389.19 ± 105.64) | 777.01 (430.59 ± 92.15) | |
| His | 70 | 249.20 | 252 | 56.61 (25.77 ± 8.04) | 54.26 (28.96 ± 7.36) | ||
| Leu + Ile | 240 | 292 | 308.40 | 288.50 | 236 | 224.68 (135.37 ± 37.20) | 250.77 (116.62 ± 30.92) |
| Met | 45 | 51 | 55.00 | 58.60 | 60 | 42.32 (22.13 ± 6.13) | 39.63 (20.58 ± 5.03) |
| Orn | 300 | 228 | 280.20 | 300 | 131.79 (59.56 ± 21.45) | 125.24 (59.14 ± 17.62) | |
| Phe | 100 | 100 | 130.20 | 118.20 | 99 | 72.15 (43.50 ± 8.89) | 78.30 (42.56 ± 9.50) |
| Pro | 350 | 401 | 362.30 | 354 | 280.47 (141.14 ± 39.85) | 250.56 (138.33 ± 33.24) | |
| Tyr | 260 | 275 | 296.30 | 287.00 | 270 | 347.85 (103.28 ± 58.74) | 231.79 (73.71 ± 33.91) |
| Val | 250 | 250 | 270.20 | 269.60 | 211 | 208.96 (115.11 ± 31.75) | 213.58 (100.71 ± 25.99) |
| ASA | 1 | 0.61 (0.11 ± 0.13) | 0.55 (0.10 ± 0.11) | ||||
| SUAC | 1.50 | 2 | 2.20 | 0.78 (0.63 ± 0.07) | 0.81 (0.60 ± 0.07) | ||
| ADO | 1.30 | 1.61 (0.46 ± 0.27) | 1.17 (0.39 ± 0.16) | ||||
| D-ADO | 0.06 | 0.02 (0.01 ± 0.01) | 0.03 (0.01 ± 0.01) | ||||
| C0 | 8 (L), 70 | 8 (L), 65 | 8.37 (L) | 62.47 | 8.20 (L), 60 | 10.42*, 71.61 (29.83 ± 10.88) | 9.18*, 49.24 (20.34 ± 6.34) |
| C2 | 5 (L), 55 | 7 (L) | 7.03 (L) | 55.45 | 49.56 | 4.24*, 35.96 (18.72 ± 7.08) | 5.64*, 43.14 (17.04 ± 6.30) |
| C3 | 5.50 | 5.20 | 5.31 | 5.45 | 7.18 | 3.96 (1.73 ± 0.78) | 4.13 (1.76 ± 0.69) |
| C3DC + C4OH | 0.30 | 0.42 | 0.49 | 0.57 | 0.20 (0.09 ± 0.03) | 0.23 (0.08 ± 0.04) | |
| C4 | 0.54 | 1.05 | 0.95 | 0.72 | 0.70 | 0.58 (0.22 ± 0.08) | 0.38 (0.18 ± 0.06) |
| C4DC + C5OH | 0.65 | 0.80 | 0.71 | 0.66 | 0.60 | 0.36 (0.18 ± 0.05) | 0.39 (0.21 ± 0.05) |
| C5 | 0.50 | 0.63 | 0.67 | 0.67 | 0.47 | 0.43 (0.20 ± 0.07) | 0.25 (0.14 ± 0.04) |
| C5:1 | 0.05 | 0.11 | 0.15 | 0.20 | 0.02 (0.01 ± 0.00) | 0.02 (0.01 ± 0.00) | |
| C5DC + C6OH | 0.20 | 0.30 | 0.34 | 0.28 | 0.30 | 0.16 (0.09 ± 0.02) | 0.17 (0.09 ± 0.02) |
| C6 | 0.23 | 0.24 | 0.27 | 0.28 | 0.15 | 0.12 (0.05 ± 0.02) | 0.12 (0.05 ± 0.02) |
| C6DC | 0.20 | 0.20 | 0.16 (0.06 ± 0.03) | 0.12 (0.05 ± 0.02) | |||
| C8 | 0.30 | 0.31 | 0.34 | 0.33 | 0.50 | 0.16 (0.07 ± 0.03) | 0.14 (0.04 ± 0.02) |
| C8:1 | 0.40 | 0.33 | 0.29 (0.11 ± 0.05) | 0.22 (0.09 ± 0.03) | |||
| C10 | 0.35 | 0.35 | 0.36 | 0.37 | 0.40 | 0.23 (0.10 ± 0.04) | 0.25 (0.10 ± 0.04) |
| C10:1 | 0.16 | 0.22 | 0.25 | 0.22 | 0.09 (0.04 ± 0.02) | 0.08 (0.04 ± 0.01) | |
| C10:2 | 0.05 | 0.11 | 0.15 | 0.12 | 0.03 (0.01 ± 0.00) | 0.03 (0.01 ± 0.00) | |
| C12 | 0.35 | 0.49 | 0.41 | 0.51 | 0.25 (0.06 ± 0.03) | 0.30 (0.07 ± 0.04) | |
| C12:1 | 0.30 | 0.35 | 0.17 (0.02 ± 0.02) | 0.25 (0.03 ± 0.03) | |||
| C14 | 0.50 | 0.63 | 0.60 | 0.62 | 0.61 | 0.42 (0.19 ± 0.07) | 0.44 (0.19 ± 0.06) |
| C14OH | 0.05 | 0.06 | 0.14 | 0.03 (0.01 ± 0.01) | 0.03 (0.01 ± 0.01) | ||
| C14:1 | 0.45 | 0.48 | 0.46 | 0.40 | 0.26 (0.06 ± 0.04) | 0.37 (0.07 ± 0.05) | |
| C14:2 | 0.06 | 0.13 | 0.04 (0.02 ± 0.01) | 0.04 (0.02 ± 0.01) | |||
| C16 | 7 | 7 | 6.92 | 6.21 | 5.57 | 4.99 (1.95 ± 0.87) | 5.94 (2.65 ± 0.90) |
| C16OH | 0.08 | 0.10 | 0.11 | 0.15 | 0.03 (0.01 ± 0.00) | 0.04 (0.01 ± 0.01) | |
| C16:1 | 0.60 | 0.53 | 0.36 (0.12 ± 0.06) | 0.43 (0.16 ± 0.08) | |||
| C16:1OH | 0.10 | 0.13 | 0.08 (0.04 ± 0.01) | 0.08 (0.04 ± 0.01) | |||
| C18 | 1.80 | 2 | 2.10 | 1.98 | 2.00 | 1.47 (0.76 ± 0.26) | 1.59 (0.83 ± 0.25) |
| C18OH | 0.05 | 0.09 | 0.08 | 0.08 | 0.02 (0.00 ± 0.01) | 0.02 (0.01 ± 0.01) | |
| C18:1 | 3 | 3 | 3.09 | 1.81 | 2.74 (1.41 ± 0.38) | 2.54 (1.27 ± 0.36) | |
| C18:1OH | 0.05 | 0.09 | 0.18 | 0.03 (0.01 ± 0.01) | 0.03 (0.01 ± 0.01) | ||
| C18:2 | 1 | 0.65 | 0.90 (0.47 ± 0.18) | 0.63 (0.28 ± 0.12) | |||
| C18:2OH | 0.15 | 0.10 | 0.09 (0.05 ± 0.02) | 0.09 (0.05 ± 0.02) | |||
| C20 | 0.08 | 0.05 (0.02 ± 0.01) | 0.06 (0.02 ± 0.01) | ||||
| C22 | 0.03 | 0.01 (0.01 ± 0.00) | 0.01 (0.01 ± 0.00) | ||||
| C24 | 0.05 | 0.03 (0.01 ± 0.01) | 0.03 (0.02 ± 0.01) | ||||
| C26 | 0.03 | 0.02 (0.01 ± 0.00) | 0.02 (0.01 ± 0.00) | ||||
| C20:0-LPC | 0.57 | 0.61 (0.32 ± 0.10) | 0.45 (0.22 ± 0.07) | ||||
| C22:0-LPC | 0.56 | 0.47 (0.24 ± 0.08) | 0.52 (0.20 ± 0.08) | ||||
| C24:0-LPC | 0.80 | 0.60 (0.36 ± 0.08) | 0.57 (0.33 ± 0.08) | ||||
| C26:0-LPC | 0.40 | 0.28 (0.16 ± 0.04) | 0.25 (0.15 ± 0.03) | ||||
Abbreviations: ASA, argininosuccinic acid; SUAC, succinylacetone; ADO, adenosine; D-ADO, 2´-deoxyadenosine; LPC, lysophosphatidylcholine; L, lower cutoff; CLIR, Collaborative Laboratory Integrated Reports; NSQAP, Newborn Screening Quality Assurance Program; amino acids are abbreviated as three-letter symbols according to the International Union of Pure and Applied Chemistry (IUPAC) nomenclature; (L), lower cutoff.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download