1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.

2. DiMasi JA, Feldman L, Seckler A, Wilson A. Trends in risks associated with new drug development: success rates for investigational drugs. Clin Pharmacol Ther. 2010; 87:272–7.

3. Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012; 22:571–84.
4. Mittal VK, Bhullar JS, Jayant K. Animal models of human colorectal cancer: current status, uses and limitations. World J Gastroenterol. 2015; 21:11854–61.

5. Evans JP, Sutton PA, Winiarski BK, Fenwick SW, Malik HZ, Vimalachandran D, et al. From mice to men: murine models of colorectal cancer for use in translational research. Crit Rev Oncol Hematol. 2016; 98:94–105.

6. Brown KM, Xue A, Mittal A, Samra JS, Smith R, Hugh TJ. Patient-derived xenograft models of colorectal cancer in pre-clinical research: a systematic review. Oncotarget. 2016; 7:66212–25.

7. Kageyama K, Ohara M, Saito K, Ozaki S, Terai M, Mastrangelo MJ, et al. Establishment of an orthotopic patient-derived xenograft mouse model using uveal melanoma hepatic metastasis. J Transl Med. 2017; 15:145.

8. Sun FX, Sasson AR, Jiang P, An Z, Gamagami R, Li L, et al. An ultra-metastatic model of human colon cancer in nude mice. Clin Exp Metastasis. 1999; 17:41–8.
9. Sewda K, Coppola D, Enkemann S, Yue B, Kim J, Lopez AS, et al. Cell-surface markers for colon adenoma and adenocarcinoma. Oncotarget. 2016; 7:17773–89.

10. Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994; 107:1183–8.

11. Fletcher RH. Carcinoembryonic antigen. Ann Intern Med. 1986; 104:66–73.

12. Diez M, Cerdan FJ, Pollan M, Maestro ML, Ortega MD, Martinez S, et al. Prognostic significance of preoperative serum CA 19.9 assay in patients with colorectal carcinoma. Anticancer Res. 1994; 14:2819–25.
13. Kemper K, Versloot M, Cameron K, Colak S, de Sousa e Melo F, de Jong JH, et al. Mutations in the Ras-Raf Axis underlie the prognostic value of CD133 in colorectal cancer. Clin Cancer Res. 2012; 18:3132–41.

14. Blaj C, Schmidt EM, Lamprecht S, Hermeking H, Jung A, Kirchner T, et al. Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Cancer Res. 2017; 77:1763–74.

15. Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009; 9:537–49.

16. Deschenes-Simard X, Kottakis F, Meloche S, Ferbeyre G. ERKs in cancer: friends or foes? Cancer Res. 2014; 74:412–9.

17. Neuzillet C, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. MEK in cancer and cancer therapy. Pharmacol Ther. 2014; 141:160–71.

18. Yao X, Sun S, Zhou X, Guo W, Zhang L. IGF-binding protein 2 is a candidate target of therapeutic potential in cancer. Tumour Biol. 2016; 37:1451–9.

19. Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016; 17:1721–30.
20. Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, et al. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov Today. 2020; 25:406–13.

21. Gan Y, Li Y, Li T, Shu G, Yin G. CCNA2 acts as a novel biomarker in regulating the growth and apoptosis of colorectal cancer. Cancer Manag Res. 2018; 10:5113–24.

22. Pocard M, Tsukui H, Salmon RJ, Dutrillaux B, Poupon MF. Efficiency of orthotopic xenograft models for human colon cancers. In Vivo. 1996; 10:463–9.
23. Seol HS, Kang HJ, Lee SI, Kim NE, Kim TI, Chun SM, et al. Development and characterization of a colon PDX model that reproduces drug responsiveness and the mutation profiles of its original tumor. Cancer Lett. 2014; 345:56–64.

24. Rosfjord E, Lucas J, Li G, Gerber HP. Advances in patient-derived tumor xenografts: from target identification to predicting clinical response rates in oncology. Biochem Pharmacol. 2014; 91:135–43.

25. Zhang Y, Zhang GL, Sun X, Cao KX, Ma C, Nan N, et al. Establishment of a murine breast tumor model by subcutaneous or orthotopic implantation. Oncol Lett. 2018; 15:6233–40.

26. Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer. 2015; 15:311–6.

27. Ayyildiz SN, Ayyildiz A. Cyanoacrylic tissue glues: Biochemical properties and their usage in urology. Turk J Urol. 2017; 43:14–24.

28. Zoccali C, Covello R, Di Francesco A, Zoccali G. A cyanoacrylate and silastic patch to reduce the risk of opening of the tumor: technical note. Eur J Surg Oncol. 2013; 39:44–5.

29. Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005; 24:2909–15.

30. Girnius N, Edwards YJ, Garlick DS, Davis RJ. The cJUN NH2-terminal kinase (JNK) signaling pathway promotes genome stability and prevents tumor initiation. Elife. 2018; 7:e36389.

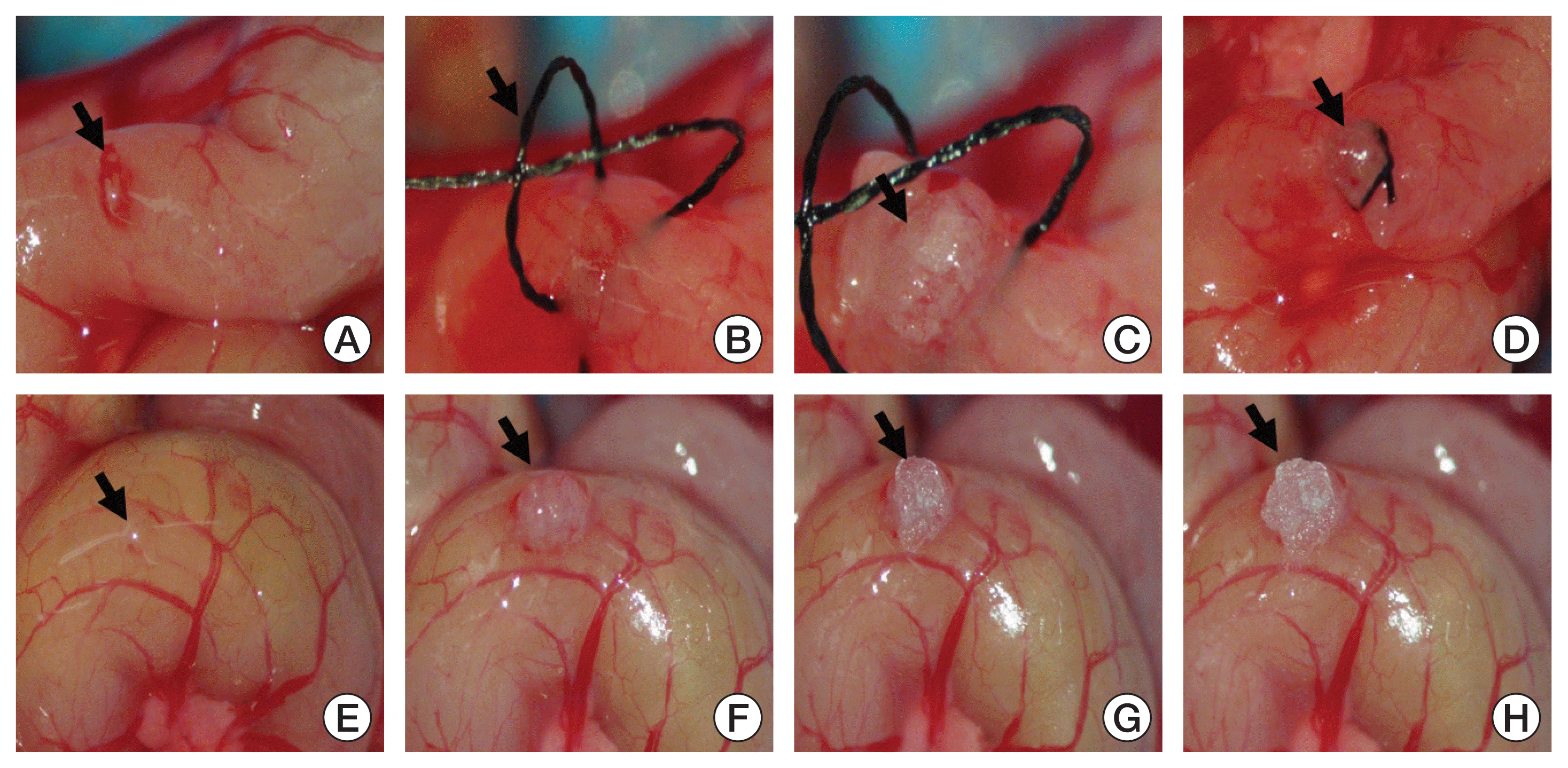
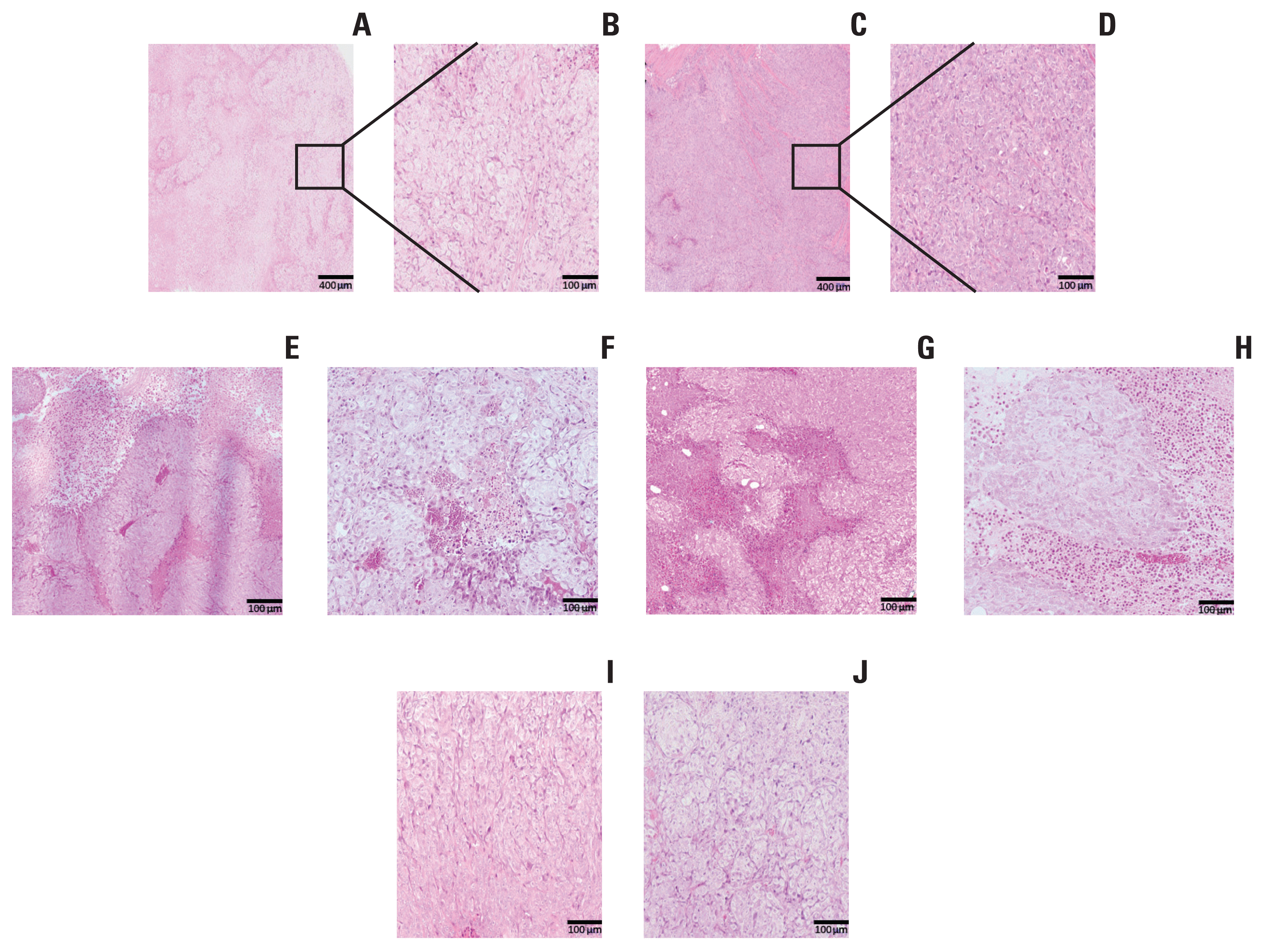
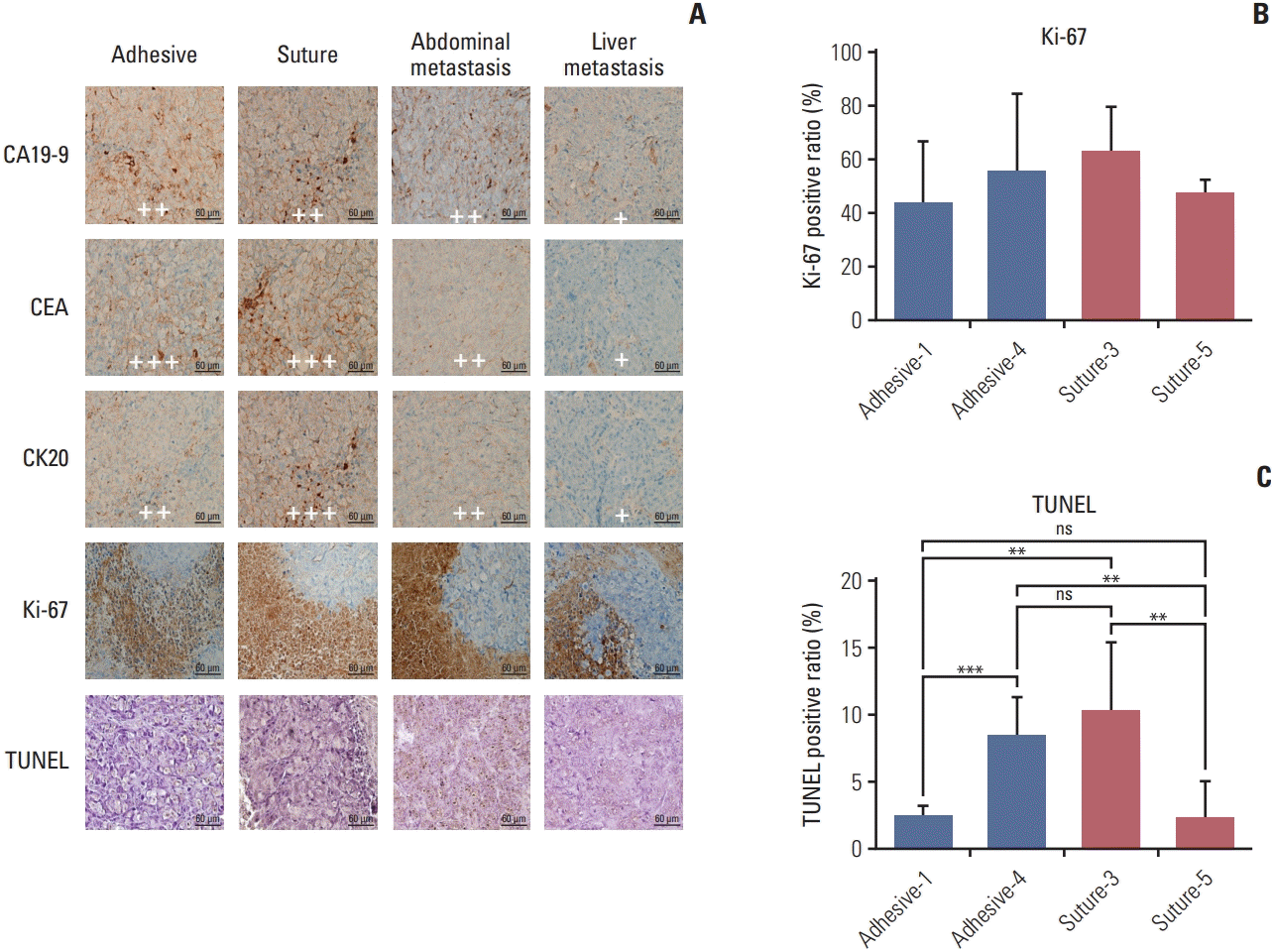
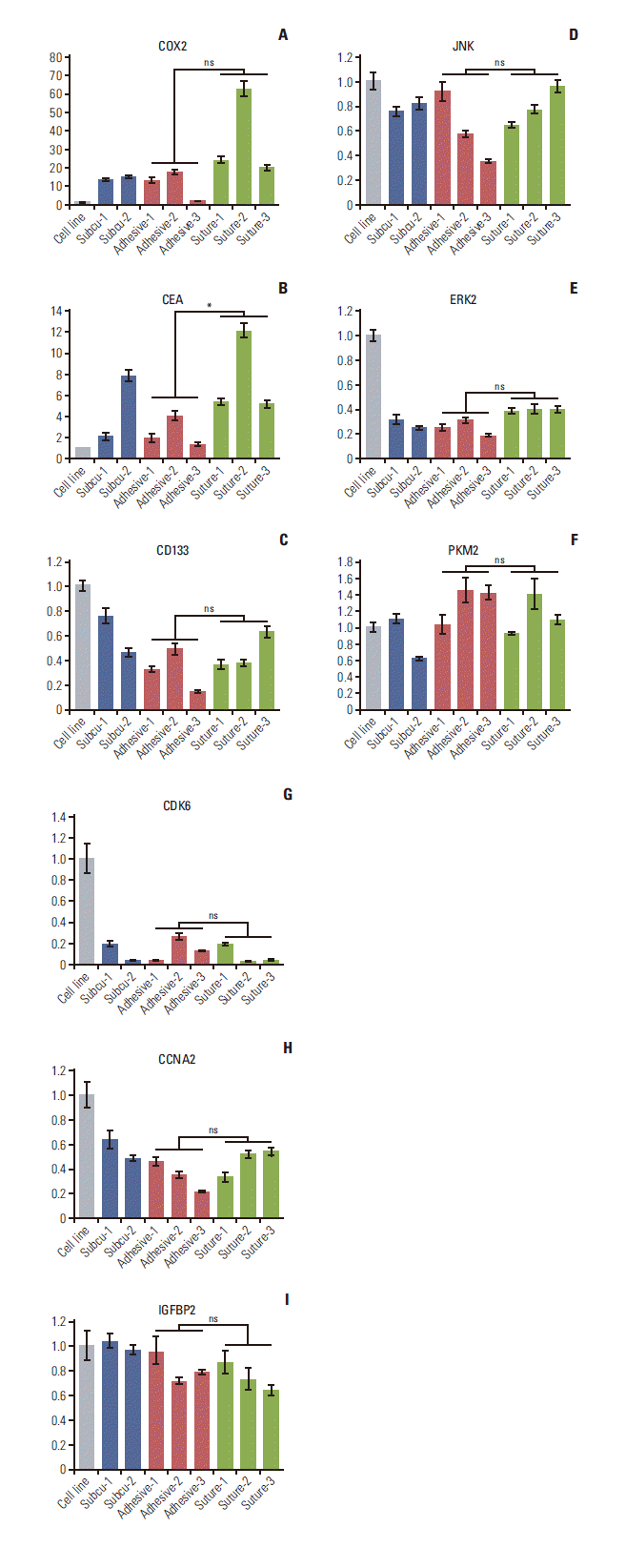




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download