1. GBD 2016 Stroke Collaborators. Global, regional, and national burden of stroke, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019; 18:439–458.
2. Saenger AK, Christenson RH. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin Chem. 2010; 56:21–33.

3. Dirnagl U, Iadecola C, Moskowitz MA. Pathobiology of ischaemic stroke: an integrated view. Trends Neurosci. 1999; 22:391–397.

4. Pfeiffer S, Anilkumar U, Chen G, Ramírez-Peinado S, Galindo-Moreno J, Muñoz-Pinedo C, et al. Analysis of BH3-only proteins upregulated in response to oxygen/glucose deprivation in cortical neurons identifies Bmf but not Noxa as potential mediator of neuronal injury. Cell Death Dis. 2014; 5.

5. Whiteley W, Tseng MC, Sandercock P. Blood biomarkers in the diagnosis of ischemic stroke: a systematic review. Stroke. 2008; 39:2902–2909.
6. Hutchison ER, Okun E, Mattson MP. The therapeutic potential of microRNAs in nervous system damage, degeneration, and repair. Neuromolecular Med. 2009; 11:153–161.

7. Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008; 33:139–147.

8. Zhao H, Wang J, Gao L, Wang R, Liu X, Gao Z, et al. MiRNA-424 protects against permanent focal cerebral ischemia injury in mice involving suppressing microglia activation. Stroke. 2013; 44:1706–1713.

9. Zeng L, He X, Wang Y, Tang Y, Zheng C, Cai H, et al. MicroRNA-210 overexpression induces angiogenesis and neurogenesis in the normal adult mouse brain. Gene Ther. 2014; 21:37–43.

10. Liu Y, Pan Q, Zhao Y, He C, Bi K, Chen Y, et al. MicroRNA-155 regulates ROS production, NO generation, apoptosis and multiple functions of human brain microvessel endothelial cells under physiological and pathological conditions. J Cell Biochem. 2015; 116:2870–2881.

11. Zhang Y, Han B, He Y, Li D, Ma X, Liu Q, et al. MicroRNA-132 attenuates neurobehavioral and neuropathological changes associated with intracerebral hemorrhage in mice. Neurochem Int. 2017; 107:182–190.

12. Fu X, Niu T, Li X. MicroRNA-126-3p attenuates intracerebral hemorrhage-induced blood-brain barrier disruption by regulating VCAM-1 expression. Front Neurosci. 2019; 13:866.

13. Yuan B, Shen H, Lin L, Su T, Zhong L, Yang Z. MicroRNA367 negatively regulates the inflammatory response of microglia by targeting IRAK4 in intracerebral hemorrhage. J Neuroinflammation. 2015; 12:206.

14. Pfeiffer S, Sánchez-Lechuga B, Donovan P, Halang L, Prehn JHM, Campos-Caro A, et al. Circulating miR-330-3p in late pregnancy is associated with pregnancy outcomes among lean women with GDM. Sci Rep. 2020; 10:908.

15. Raoof R, Jimenez-Mateos EM, Bauer S, Tackenberg B, Rosenow F, Lang J, et al. Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci Rep. 2017; 7:3328.

16. Bacon S, Engelbrecht B, Schmid J, Pfeiffer S, Gallagher R, McCarthy A, et al. MicroRNA-224 is readily detectable in urine of individuals with diabetes mellitus and is a potential indicator of beta-cell demise. Genes (Basel). 2015; 6:399–416.
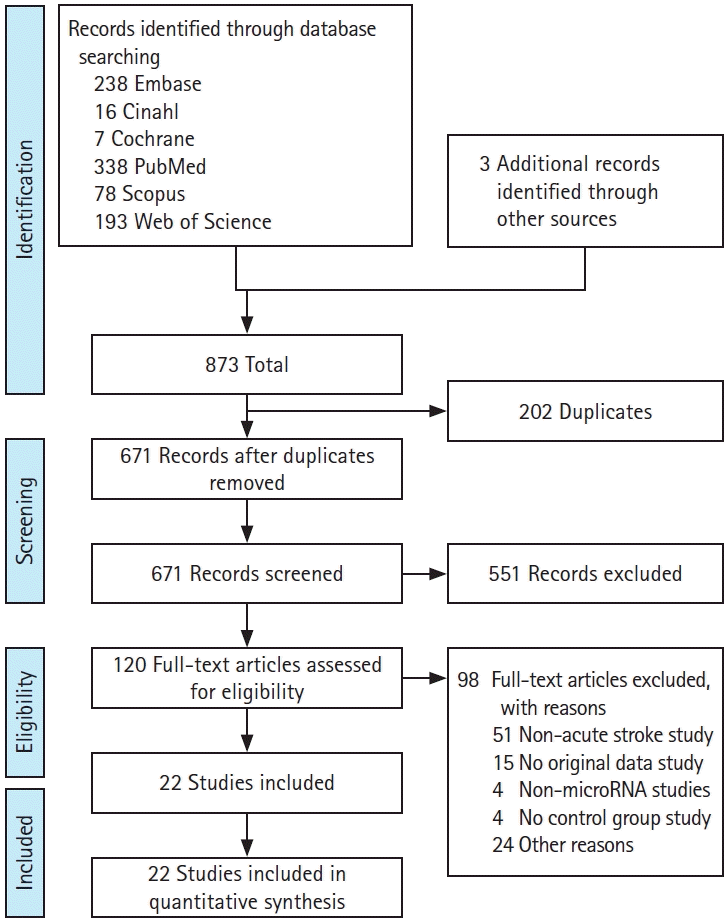
17. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009; 6:e1000097.

18. Lo CK, Mertz D, Loeb M. Newcastle-Ottawa Scale: comparing reviewers’ to authors’ assessments. BMC Med Res Methodol. 2014; 14:45.

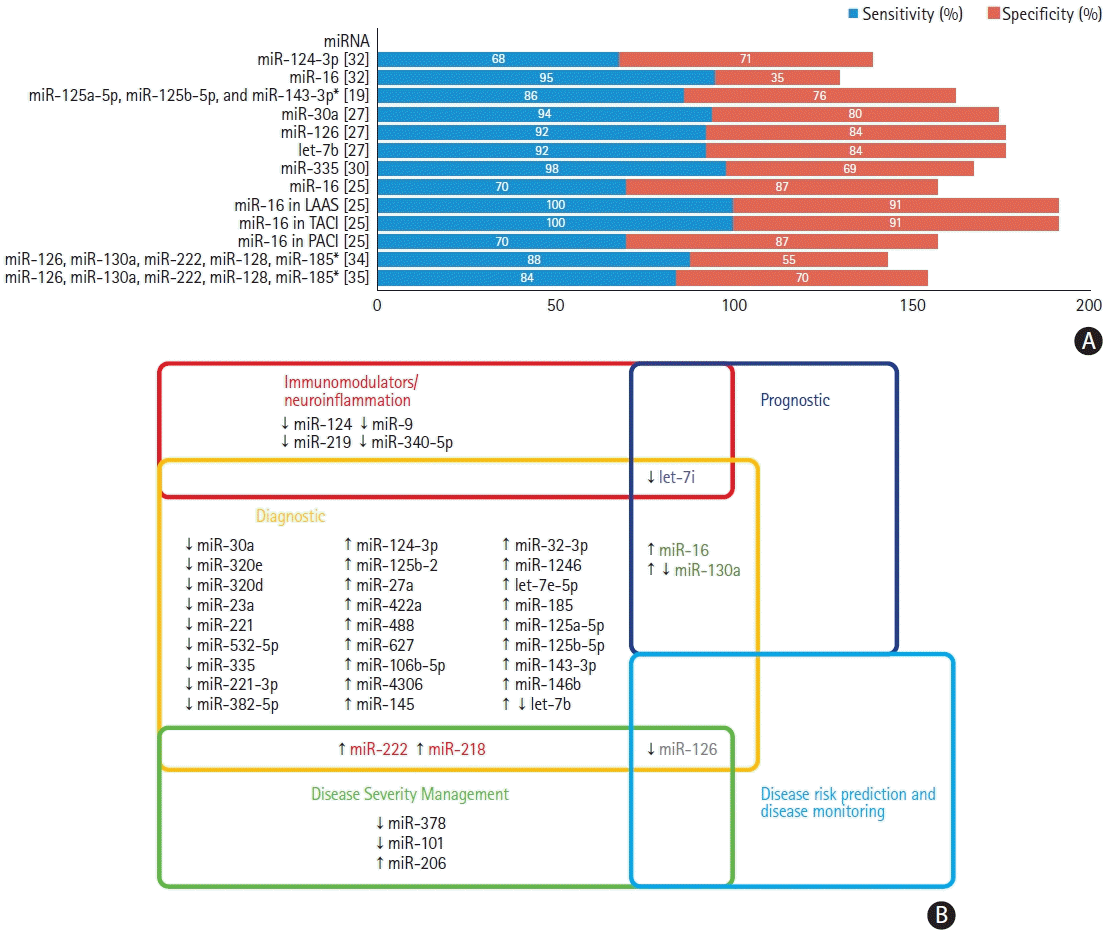
19. Tiedt S, Prestel M, Malik R, Schieferdecker N, Duering M, Kautzky V, et al. RNA-Seq identifies circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res. 2017; 121:970–980.

20. Sepramaniam S, Tan JR, Tan KS, DeSilva DA, Tavintharan S, Woon FP, et al. Circulating microRNAs as biomarkers of acute stroke. Int J Mol Sci. 2014; 15:1418–1432.

21. Xiang W, Tian C, Lin J, Wu X, Pang G, Zhou L, et al. Plasma let-7i and miR-15a expression are associated with the effect of recombinant tissue plasminogen activator treatment in acute ischemic stroke patients. Thromb Res. 2017; 158:121–125.

22. Li G, Ma Q, Wang R, Fan Z, Tao Z, Liu P, et al. Diagnostic and immunosuppressive potential of elevated Mir-424 levels in circulating immune cells of ischemic stroke patients. Aging Dis. 2018; 9:172–181.

23. Ma Q, Li G, Tao Z, Wang J, Wang R, Liu P, et al. Blood microRNA-93 as an indicator for diagnosis and prediction of functional recovery of acute stroke patients. J Clin Neurosci. 2019; 62:121–127.

24. Yoo H, Kim J, Lee AR, Lee JM, Kim OJ, Kim JK, et al. Alteration of microRNA 340-5p and arginase-1 expression in peripheral blood cells during acute ischemic stroke. Mol Neurobiol. 2019; 56:3211–3221.

25. Tian C, Li Z, Yang Z, Huang Q, Liu J, Hong B. Plasma microRNA-16 is a biomarker for diagnosis, stratification, and prognosis of hyperacute cerebral infarction. PLoS One. 2016; 11:e0166688.

26. Liu Y, Zhang J, Han R, Liu H, Sun D, Liu X. Downregulation of serum brain specific microRNA is associated with inflammation and infarct volume in acute ischemic stroke. J Clin Neurosci. 2015; 22:291–295.

27. Long G, Wang F, Li H, Yin Z, Sandip C, Lou Y, et al. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013; 13:178.

28. Huang S, Lv Z, Guo Y, Li L, Zhang Y, Zhou L, et al. Identification of blood let-7e-5p as a biomarker for ischemic stroke. PLoS One. 2016; 11:e0163951.

29. Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol. 2015; 35:433–447.

30. Zhao B, Zhu Z, Hao J, Wan Z, Guo X. Decreased plasma miR-335 expression in patients with acute ischemic stroke and its association with calmodulin expression. J Int Med Res. 2016; 44:1331–1338.

31. Adams HP Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993; 24:35–41.

32. Leung LY, Chan CP, Leung YK, Jiang HL, Abrigo JM, Wang de F, et al. Comparison of miR-124-3p and miR-16 for early diagnosis of hemorrhagic and ischemic stroke. Clin Chim Acta. 2014; 433:139–144.

33. Wang W, Sun G, Zhang L, Shi L, Zeng Y. Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J Stroke Cerebrovasc Dis. 2014; 23:2607–2613.

34. Jin F, Xing J. Circulating pro-angiogenic and anti-angiogenic microRNA expressions in patients with acute ischemic stroke and their association with disease severity. Neurol Sci. 2017; 38:2015–2023.

35. Jin F, Xing J. Circulating miR-126 and miR-130a levels correlate with lower disease risk, disease severity, and reduced inflammatory cytokine levels in acute ischemic stroke patients. Neurol Sci. 2018; 39:1757–1765.

36. Wang MD, Wang Y, Xia YP, Dai JW, Gao L, Wang SQ, et al. High serum miR-130a levels are associated with severe perihematomal edema and predict adverse outcome in acute ICH. Mol Neurobiol. 2016; 53:1310–1321.

37. Chen Z, Wang K, Huang J, Zheng G, Lv Y, Luo N, et al. Upregulated serum miR-146b serves as a biomarker for acute ischemic stroke. Cell Physiol Biochem. 2018; 45:397–405.

38. Jia L, Hao F, Wang W, Qu Y. Circulating miR-145 is associated with plasma high-sensitivity C-reactive protein in acute ischemic stroke patients. Cell Biochem Funct. 2015; 33:314–319.

39. Wang Y, Ma Z, Kan P, Zhang B. The diagnostic value of serum miRNA-221-3p, miRNA-382-5p, and miRNA-4271 in ischemic stroke. J Stroke Cerebrovasc Dis. 2017; 26:1055–1060.

40. Zhou J, Chen L, Chen B, Huang S, Zeng C, Wu H, et al. Increased serum exosomal miR-134 expression in the acute ischemic stroke patients. BMC Neurol. 2018; 18:198.

41. Jickling GC, Ander BP, Shroff N, Orantia M, Stamova B, Dykstra-Aiello C, et al. Leukocyte response is regulated by microRNA let7i in patients with acute ischemic stroke. Neurology. 2016; 87:2198–2205.

42. Yang X, Tang X, Sun P, Shi Y, Liu K, Hassan SH, et al. MicroRNA-15a/16-1 antagomir ameliorates ischemic brain injury in experimental stroke. Stroke. 2017; 48:1941–1947.

43. Ma M, Ma Y, Yi X, Guo R, Zhu W, Fan X, et al. Intranasal delivery of transforming growth factor-beta1 in mice after stroke reduces infarct volume and increases neurogenesis in the subventricular zone. BMC Neurosci. 2008; 9:117.

44. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007; 129:1401–1414.

45. Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res. 2013; 112:335–346.

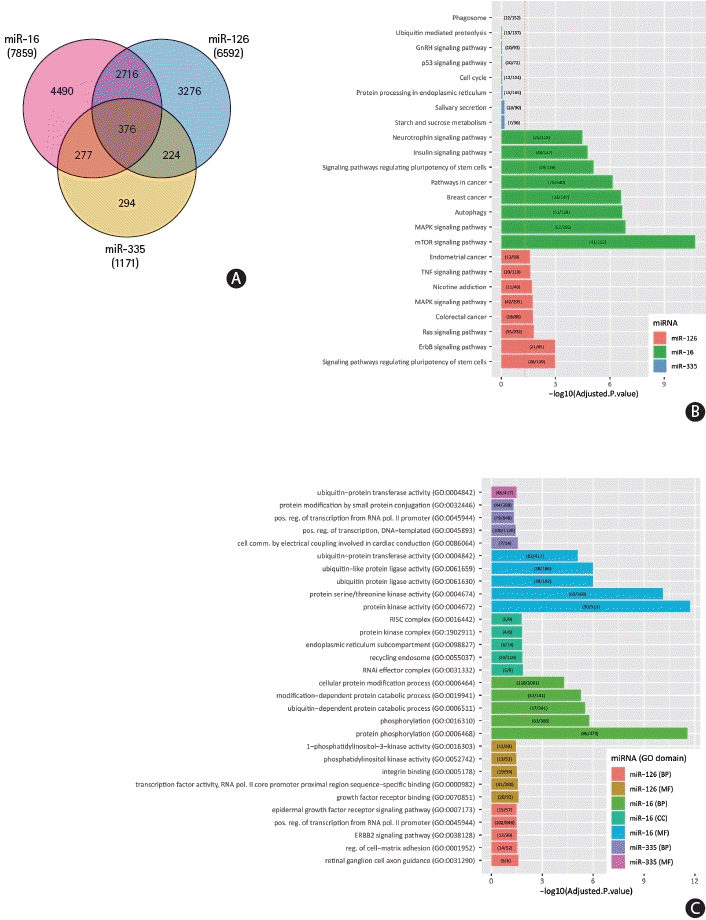
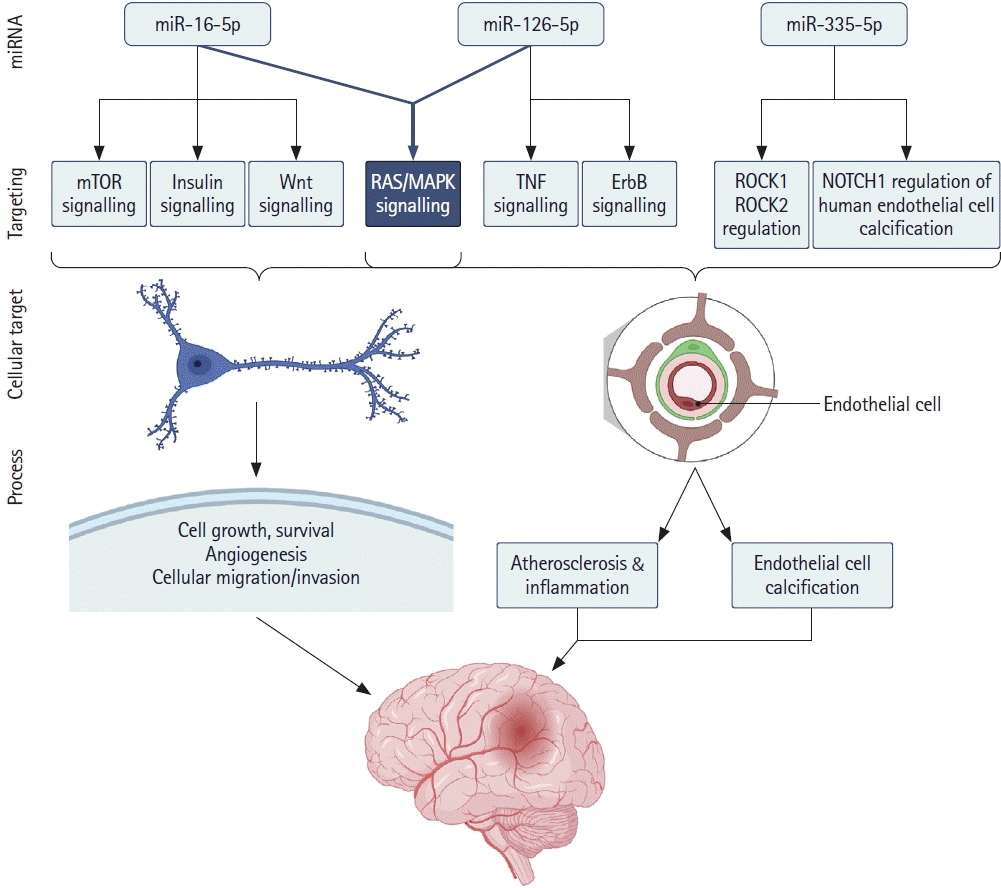
46. Maiese K. Cutting through the complexities of mTOR for the treatment of stroke. Curr Neurovasc Res. 2014; 11:177–186.

47. Pomytkin I, Krasil’nikova I, Bakaeva Z, Surin A, Pinelis V. Excitotoxic glutamate causes neuronal insulin resistance by inhibiting insulin receptor/Akt/mTOR pathway. Mol Brain. 2019; 12:112.

48. van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren-Rietdijk AM, et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med. 2009; 13:1577–1585.

49. Kim KJ, Cho CS, Kim WU. Role of placenta growth factor in cancer and inflammation. Exp Mol Med. 2012; 44:10–19.

50. Jakob P, Doerries C, Briand S, Mocharla P, Kränkel N, Besler C, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012; 126:2962–2975.
51. Tang ST, Wang F, Shao M, Wang Y, Zhu HQ. MicroRNA-126 suppresses inflammation in endothelial cells under hyperglycemic condition by targeting HMGB1. Vascul Pharmacol. 2017; 88:48–55.

52. Qu WS, Tian DS, Guo ZB, Fang J, Zhang Q, Yu ZY, et al. Inhibition of EGFR/MAPK signaling reduces microglial inflammatory response and the associated secondary damage in rats after spinal cord injury. J Neuroinflammation. 2012; 9:178.

53. Si W, Ye S, Ren Z, Liu X, Wu Z, Li Y, et al. miR‑335 promotes stress granule formation to inhibit apoptosis by targeting ROCK2 in acute ischemic stroke. Int J Mol Med. 2019; 43:1452–1466.

54. Gošev I, Zeljko M, Đurić Ž, Nikolić I, Gošev M, Ivčević S, et al. Epigenome alterations in aortic valve stenosis and its related left ventricular hypertrophy. Clin Epigenetics. 2017; 9:106.

55. Wang Y, Wang MD, Xia YP, Gao Y, Zhu YY, Chen SC, et al. MicroRNA-130a regulates cerebral ischemia-induced blood-brain barrier permeability by targeting Homeobox A5. FASEB J. 2018; 32:935–944.

56. Huang RS, Gamazon ER, Ziliak D, Wen Y, Im HK, Zhang W, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011; 8:692–701.

57. Mompeón A, Ortega-Paz L, Vidal-Gómez X, Costa TJ, Pérez-Cremades D, Garcia-Blas S, et al. Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Sci Rep. 2020; 10:5373.
58. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010; 50:298–301.

59. Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015; 61:1333–1342.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download