Introduction
Materials and Methods
1. Study patients
2. Targeted DNA sequencing and bioinformatics
3. Statistical analysis
Results
1. Prevalence and characteristics of PIK3CA and AKT mutations
Table 1.
| Gene | Mutation type | Occurrence | Status | Hot-spot mutation | Frequency (%) |
|---|---|---|---|---|---|
| AKT1 | AKT1_E17K | 17 | Tumor | Yes | 3.4 |
| AKT1_C77F | 1 | Tumor | No | 0.2 | |
| PIK3CAa) | PIK3CA_H1047R | 121 | Tumor | Yes | 23.9 |
| PIK3CA_E545K | 50 | Tumor | Yes | 9.9 | |
| PIK3CA_H1047L | 19 | Tumor | Yes | 37.5 | |
| PIK3CA_E542K | 14 | Tumor | Yes | 27.6 | |
| PIK3CA_N345K | 8 | Tumor | No | 1.6 | |
| PIK3CA_E726K | 7 | Tumor | No | 1.4 | |
| PIK3CA_C420R | 5 | Tumor | No | 1.0 | |
| PIK3CA_G118D | 3 | Tumor | No | 0.6 | |
| PIK3CA_E365K | 2 | Tumor | No | 0.4 | |
| PIK3CA_P471L | 2 | Tumor | No | 0.4 | |
| PIK3CA_Q546K | 2 | Tumor | No | 0.4 | |
| PIK3CA_T1025A | 2 | Tumor | No | 0.4 | |
| PIK3CA_G1049R | 2 | Tumor | No | 0.4 |
a) 36 additional PIK3CA variants (tumor status) were identified with 1 occurrence: PIK3CA_C420DEL, PIK3CA_D454DEL, PIK3CA_D454N, PIK3CA_D527N, PIK3CA_D830H, PIK3CA_E103DEL, PIK3CA_E418K, PIK3CA_E545D, PIK3CA_E545Q, PIK3CA_E600K, PIK3CA_E674Q, PIK3CA_E722Q, PIK3CA_E798K, PIK3CA_E81K, PIK3CA_E970K, PIK3CA_E9DE, PIK3CA_G364R, PIK3CA_H1065L, PIK3CA_HC491DEL, PIK3CA_I102F, PIK3CA_K111E, PIK3CA_K485N, PIK3CA_KI111N, PIK3CA_L422W, PIK3CA_M16I, PIK3CA_N1044I, PIK3CA_N1068INS, PIK3CA_N345I, PIK3CA_p.Cys472_Leu473insPheGlu, PIK3CA_P104DEL, PIK3CA_P449DEL, PIK3CA_Q546E, PIK3CA_Q75E, PIK3CA_R115L, PIK3CA_T1052K PIK3CA_V101DEL.
Table 2.
| Characteristic |
AKT1 mutation |
p-value |
PIK3CA mutation |
p-value |
PIK3CA/AKT1 mutation burdens |
p-value | ||
|---|---|---|---|---|---|---|---|---|
| MT (n=18) | Hotspot MT (n=200) | Non-hotspot MT (n=36) | 2MTs (n=39) | 1MT (n=211) | ||||
| Age (yr) | 50.9±9.3 | 0.72a) | 50.8±10.1 | 51.2±8.3 | 0.24b) | 50.6±9.4 | 50.9±9.9 | 0.20c) |
| Menopausal status | 0.60a) | 0.92b) | 0.92c) | |||||
| Premenopausal | 9 (50.0) | 88 (44.0) | 17 (47.2) | 0.720d) | 17 (43.6) | 95 (45.0) | 0.856e) | |
| Postmenopausal | 9 (50.0) | 112 (56.0) | 19 (52.8) | 22 (56.4) | 116 (55.0) | |||
| Breast cancer family history | 0.53a) | 0.29b) | 0.79c) | |||||
| Yes | 1 (5.6) | 4 (2.0) | 2 (5.6) | 0.225d) | 0 | 7 (3.3) | 0.600e) | |
| No | 17 (94.4) | 198 (98.9) | 34 (94.4) | 39 (100) | 204 (96.7) | |||
| Pathological type | 0.59a) | 0.68b) | 0.98c) | |||||
| DCIS | 0 | 1 (0.5) | 1 (2.8) | 0.382d) | 0 | 2 (0.9) | 1.0e) | |
| IDC | 17 (94.4) | 192 (96.0) | 34 (94.4) | 38 (97.4) | 201 (95.3) | |||
| Other IBC | 1 (5.6) | 7 (3.5) | 1 (2.8) | 1 (2.6) | 8 (3.8) | |||
| Tumor grade | 0.06a) | 0.14b) | 0.04c) | |||||
| 1 | 1 (5.6) | 10 (5.0) | 1 (2.8) | 0.149d) | 3 (7.7) | 8 (3.8) | 0.066e) | |
| 2 | 10 (55.6) | 67 (33.5) | 9 (25.0) | 19 (48.7) | 64 (30.3) | |||
| 3 | 7 (38.9) | 108 (54.0) | 26 (72.2) | 15 (38.5) | 126 (59.7) | |||
| Unknown | 0 | 15 (7.5) | 0 | 2 (5.1) | 13 (6.2) | |||
| T stage | 0.08a) | 0.64b) | 0.59c) | |||||
| Tis/T1 | 9 (50.0) | 70 (35.0) | 10 (27.8) | 0.395d) | 11 (28.2) | 75 (35.5) | 0.495e) | |
| T2 | 7 (38.9) | 118 (59.0) | 24 (66.7) | 26 (66.7) | 122 (57.8) | |||
| T3 | 0 | 7 (3.5) | 0 | 0 | 7 (3.3) | |||
| T4 | 2 (11.1) | 5 (2.5) | 2 (5.6) | 2 (5.1) | 7 (3.3) | |||
| Tx | 0 | 0 | 0 | 0 | 0 | |||
| Lymph node involvement | 0.90a) | 0.46b) | 0.56c) | |||||
| N0 | 9 (50.0) | 95 (47.5) | 13 (36.1) | 0.436d) | 15 (38.5) | 99 (46.9) | 0.713e) | |
| N1 | 7 (38.9) | 66 (33.0) | 17 (47.2) | 17 (43.6) | 72 (34.1) | |||
| N2 | 1 (5.6) | 25 (12.5) | 4 (11.1) | 4 (10.3) | 26 (12.3) | |||
| N3 | 1 (5.6) | 14 (7.0) | 2 (5.6) | 3 (7.7) | 14 (6.6) | |||
| Metastasis | 0.17a) | 0.13b) | 0.598c) | |||||
| M0 | 17 (94.4) | 200 (100) | 36 (100) | 39 (100) | 210 (99.5) | |||
| M1 | 1 (5.6) | 0 | 0 | 0 | 1 (0.5) | |||
| Molecular subtypes | 0.001a) | 0.003b) | < 0.001c) | |||||
| Luminal A | 6 (33.3) | 25 (12.5) | 4 (11.1) | 0.169d) | 10 (25.6) | 23 (10.9) | 0.140e) | |
| Luminal B1 | 12 (66.7) | 110 (55.0) | 13 (36.1) | 21 (53.8) | 112 (53.1) | |||
| Luminal B2 | 0 | 28 (14.0) | 8 (22.2) | 3 (7.7) | 33 (15.6) | |||
| HER2+ | 0 | 21 (10.5) | 7 (19.4) | 3 (7.7) | 25 (11.8) | |||
| TN | 0 | 16 (8.0) | 4 (11.1) | 2 (5.1) | 18 (8.5) | |||
| ER | 0.001a) | < 0.001b) | < 0.001c) | |||||
| Positive | 18 (100) | 158 (79.0) | 25 (69.4) | 0.206d) | 34 (87.2) | 163 (77.3) | 0.163e) | |
| Negative | 0 | 42 (21.0) | 11 (30.6) | 5 (12.8) | 48 (22.7) | |||
| PR | 0.001a) | 0.013b) | 0.005c) | |||||
| Positive | 18 (100) | 147 (73.5) | 21 (68.3) | 0.064d) | 30 (76.9) | 152 (72.0) | 0.529e) | |
| Negative | 0 | 53 (26.5) | 15 (31.7) | 9 (23.1) | 59 (28.0) | |||
| HER2 | 0.003a) | 0.039b) | 0.021c) | |||||
| Positive | 0 | 49 (24.5) | 15 (41.7) | 0.033d) | 6 (15.4) | 58 (27.5) | 0.112e) | |
| Negative | 18 (100) | 151 (75.5) | 21 (58.3) | 33 (84.6) | 153 (72.5) | |||
| Ki67 | 0.001a) | 0.62b) | 0.031c) | |||||
| High | 10 (55.6) | 163 (81.5) | 30 (83.3) | 0.793d) | 27 (69.2) | 175 (82.9) | 0.046e) | |
| Low | 8 (44.4) | 37 (19.5) | 6 (16.7) | 12 (30.8) | 36 (17.1) | |||
Value are presented as number (%). MT, mutation; 2MTs, 2 or 3 mutations; 1MT, 1 mutation; DCIS, ductal carcinoma in situ; IDC, invasive ductal carcinoma; IBC, invasive breast cancer; HER2, human epidermal growth factor receptor 2; TN, triple negative subtype; Luminal B1, luminal B-HER2 negative subtype; Luminal B2, luminal B2-HER2 positive subtype; HER2+, HER2 positive subtype; ER, estrogen receptor; PR, progesterone receptor.
1) AKT1 mutation
2) PIK3CA mutation
3) PIK3CA/AKT1 mutation
2. Association of PIK3CA and AKT1 mutations with survival
1) Total patients (296 cases)
Fig. 1.
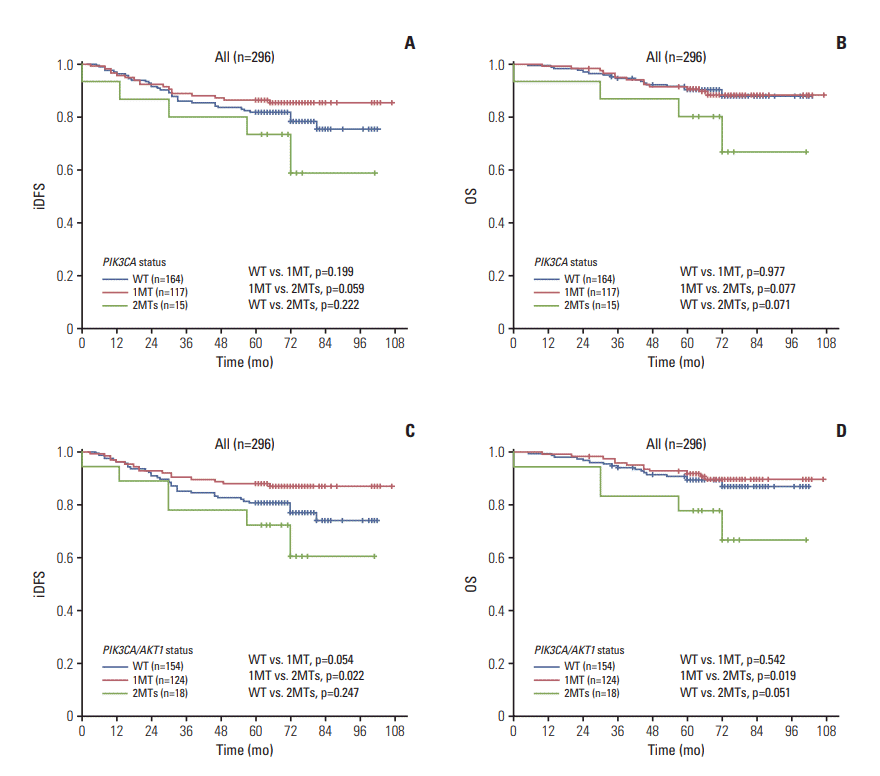
Table 3.
|
PIK3CA mutation |
PIK3CA/AKT1 mutation |
|||||||
|---|---|---|---|---|---|---|---|---|
|
iDFS |
OS |
iDFS |
OS |
|||||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Total (n=296) | ||||||||
| 2MTs | 1a) | 0.119 | 1a) | 0.053 | 1b) | 0.056 | 1b) | 0.008 |
| 1MT | 0.344 (0.119-0.994) | 0.049 | 0.222 (0.063-0.787) | 0.020 | 0.269 (0.098-0.742) | 0.011 | 0.151 (0.045-0.510) | 0.002 |
| WT | 0.500 (0.177-1.414) | 0.191 | 0.221 (0.061-0.799) | 0.021 | 0.452 (0.169-1.211) | 0.114 | 0.181 (0.053-0.612) | 0.006 |
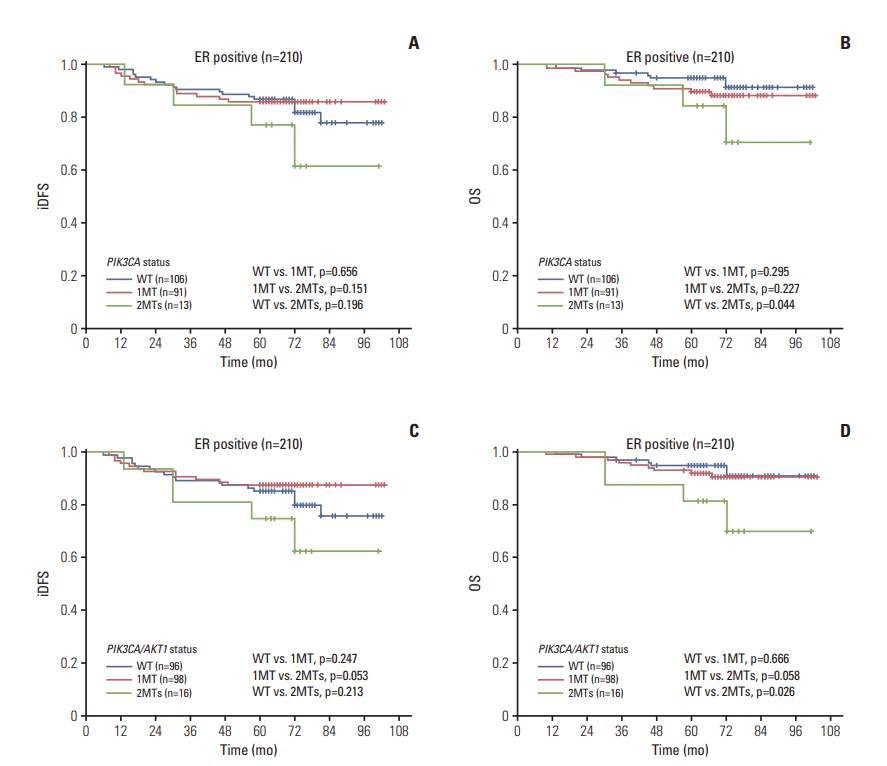
| ER+ (n=210) | 1c) | 0.402 | 1c) | 0.165 | 1d) | 0.125 | 1d) | 0.053 |
| 1MT | 0.444 (0.133-1.482) | 0.187 | 0.333 (0.073-1.518) | 0.155 | 0.322 (0.104-1.003) | 0.051 | 0.195 (0.046-0.820) | 0.026 |
| WT | 0.588 (0.180-1.921) | 0.379 | 0.203 (0.039-1.052) | 0.058 | 0.537 (0.178-1.624) | 0.271 | 0.167 (0.036-0.774) | 0.022 |
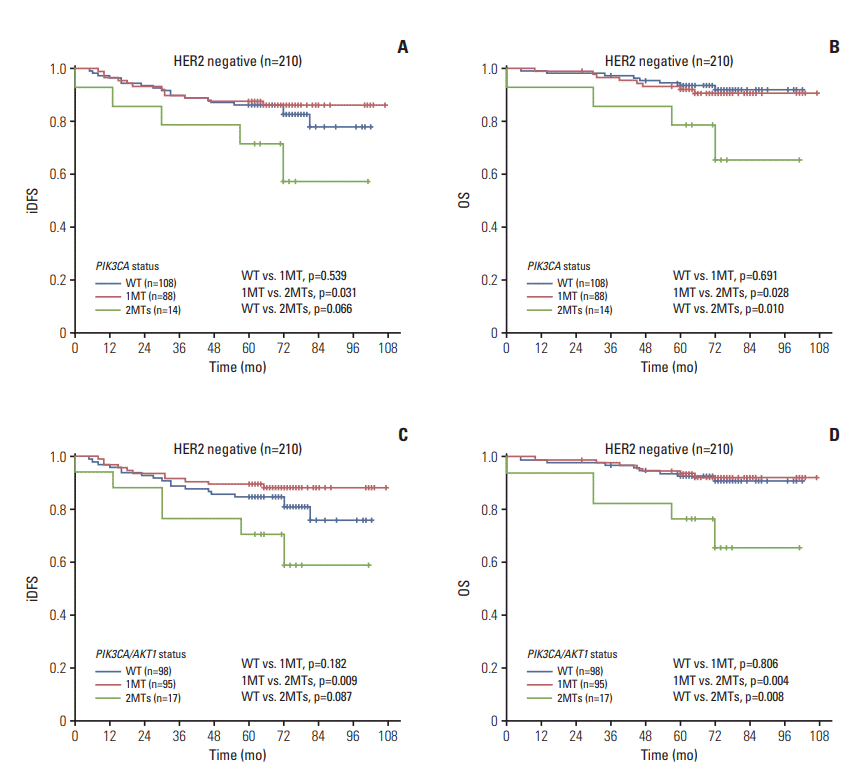
| HER2– (n=210) | 1e) | 0.105 | 1e) | 0.056 | 1f) | 0.038 | 1f) | 0.017 |
| 1MT | 0.304 (0.100-0.921) | 0.035 | 0.246 (0.062-0.972) | 0.045 | 0.246 (0.084-0.721) | 0.011 | 0.166 (0.044-0.636) | 0.009 |
| WT | 0.368 (0.121-1.116) | 0.077 | 0.171 (0.039-0.741) | 0.018 | 0.376 (0.131-1.079) | 0.069 | 0.161 (0.040-0.643) | 0.010 |
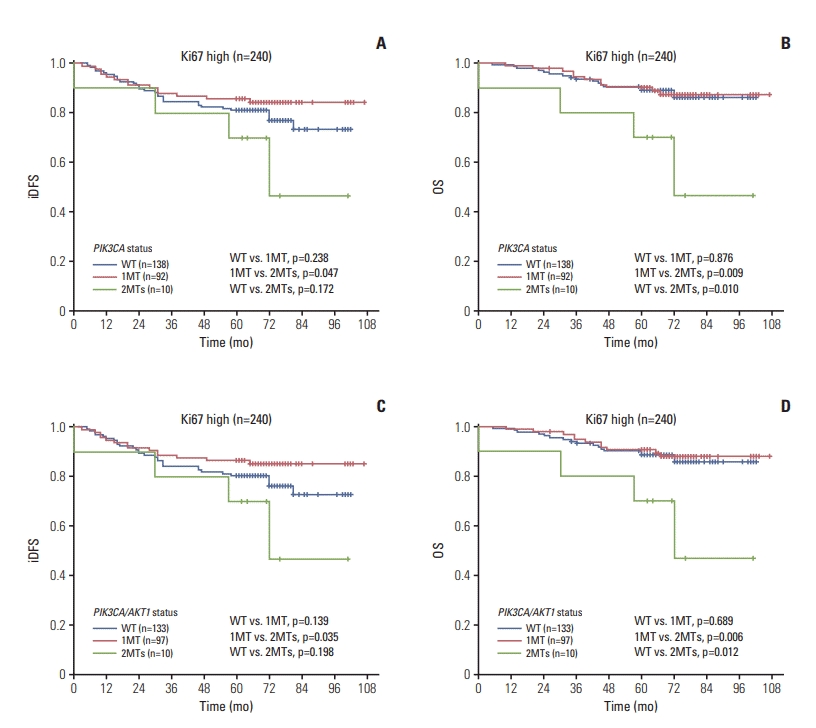
| Ki67 high (n=210) | 1g) | 0.027 | 1g) | 0.003 | 1h) | 0.022 | 1h) | 0.002 |
| 1MT | 0.198 (0.059-0.662) | 0.009 | 0.096 (0.025-0.372) | 0.001 | 0.191 (0.057-0.638) | 0.007 | 0.093 (0.024-0.362) | 0.001 |
| WT | 0.309 (0.094-1.014) | 0.053 | 0.114 (0.029-0.446) | 0.002 | 0.308 (0.094-1.015) | 0.053 | 0.163 (0.029-0.447) | 0.002 |
a) Adjusted by AKT1 mutation, age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), Ki67, histological grade, surgery, radiotherapy, chemotherapy, endocrine therapy, and target therapy,
b) Adjusted by age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, ER, PR, HER2, Ki67, histological grade, surgery, radiotherapy, chemotherapy, endocrine therapy, and target therapy,
c) Adjusted by AKT1 mutation, age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, PR, HER2, Ki67, histological grade, surgery, radiotherapy, chemotherapy, endocrine therapy, and target therapy,
d) Adjusted by age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, PR, HER2, Ki67, histological grade, surgery, radiotherapy, chemotherapy, endocrine therapy, and target therapy,
e) Adjusted by AKT1 mutation, age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, ER, PR, Ki67, histological grade, surgery, radiotherapy, chemotherapy, and endocrine therapy,
f) Adjusted by age at diagnosis, menopause at diagnosis, tumor size, lymph node metastasis, ER, PR, Ki67, histological grade, surgery, radiotherapy, chemotherapy, and endocrine therapy,
2) ER-positive patients (210 cases)
Fig. 2.
3) HER2-negative patients (210 cases)
Fig. 3.
4) Patients with high expression of Ki67 (240 cases)
Fig. 4.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download