Abstract
Purpose
MicroRNAs (miRNAs) regulate various cellular functions, including development, cell proliferation, apoptosis, and tumorigenesis. Different signatures associated with various tissue types, diagnosis, progression, prognosis, staging, and treatment response have been identified by miRNA expression profiling of human tumors. miRNAs function as oncogenes or as tumor suppressors. The relationship between gastric cancer and miRNA garnered attention due to the high incidence of gastric cancer in Asian countries. miR-222/221 expression increases in gastric tumor tissues. The oncogenic effect of miR-222/221 was previously determined in functional studies and xenograft models. In this study, transgenic mice over-expressing miR-222/221 were generated to confirm the effect of miR-222/221 on gastric carcinogenesis.
Materials and Methods
At 6 weeks of age, 65 transgenic mice and 53 wild-type mice were given drinking water containing N-nitroso-N-methylurea (MNU) for 5 alternating weeks to induce gastric cancer. The mice were euthanized at 36 weeks of age and histologic analysis was performed.
Results
Hyperplasia was observed in 3.77% of the wild-type mice and in 18.46% of the transgenic mice (p=0.020). Adenoma was observed in 20.75% of the wild-type mice and 26.15% of the transgenic mice (p=0.522). Carcinoma was observed in 32.08% of the wild-type mice and 41.54% of the transgenic mice (p=0.341). The frequency of hyperplasia, adenoma, and carcinoma was higher in transgenic mice, but the difference was statistically significant only in hyperplasia.
MicroRNAs (miRNAs) are small non-coding RNAs of about 22 nucleotides in length; they function as post-transcriptional regulators by base-pairing with the complementary sites at the 3′-untranslated region of mRNA. Since miRNA was identified, many studies of small regulators have been published [1]. miRNAs can control the stability of target mRNA and the efficiency of translation [2]. The latest version of the miRBase database (Release 18) includes 1,921 miRNAs from humans [3]. Bioinformatic analyses predict that miRNAs may regulate almost all of the human protein-coding genes [4]. Each miRNA is predicted to suppress over hundreds of target genes by target prediction programs. In other words, each target mRNA can be repressed by different miRNAs. Since the importance of miRNAs was magnified, many efforts have been made to identify miRNA target genes for individual miRNA using computational and experimental approaches.
Until now, various functions of miRNAs have been suggested, ranging from development to the life span of many diseases, especially for cancer [5]. Many targets of miRNAs are also related to human diseases, especially to cancer. For instance, cell cycle regulators, tumor suppressor genes, oncogenes, growth factors, and various genes regulating cell growth are targets of miRNAs that relate to cancer. These findings suggest that aberrant alterations of miRNA levels are linked to human malignancy [6]. Recent studies showed that, by regulating the expression of oncogenes and tumor suppressor genes, miRNAs may play critical roles in maintaining cellular homeostasis. Based on the broad scope of the target genes regulated by miRNAs, it is important to determine the role of various miRNAs and incorporate these functions into cellular signaling pathways in order to apply these findings to treatment of diseases.
Gastric cancer is the second most common cause of death from cancer worldwide, especially in many Asian countries, including China, Japan, and Korea. It is responsible for about 10% of tumor-related deaths worldwide [7]. Gastric cancer received international attention because of its high incidence and mortality rates in Eastern Asia [8]. Although the level of cell cycle regulators is known to be abnormal in gastric cancer, the molecular pathology of gastric cancer is not yet completely understood [9]. Because of the importance of gastric cancer in Eastern Asia, many groups studied the relation between microRNAs and human gastric malignancy. Differential expression of microRNAs in gastric cancer has been recently shown [10]. Furthermore, functional studies of microRNAs in gastric cancer have been reported by many groups [11,12]. In a previous study, our group investigated human miRNA profiles in gastric carcinoma compared to matched normal gastric tissue using miRNA microarray [13]. The miR-222/221 cluster, one of the most upregulated clusters, was selected as a candidate oncogene miRNA in gastric carcinoma. From this finding, we speculated that miR-222/221 overexpression in transgenic mice would promote gastric carcinogenesis by suppressing the target genes of miR-222/221, p27, and the CDK inhibitor, thereby helping cancer cells to overcome the cell-cycle checkpoint [13].
In the present study, a miR-222/221 transgenic mouse model was generated to provide further evidence supporting the carcinogenic effect of miR-222/221. N-methyl-N-nitrosourea (MNU), an alkylating agent that induces adenomas and adenocarcinomas in the stomach, was administered to the mice to induce gastric carcinogenesis [14,15]. The goals of the present study were to clarify the role of miR-222/221 in gastric carcinogenesis not only in vitro, but also in vivo by assessing the phenotype of the generated transgenic mice and by validating the suppressive effect of miR-222/221 on target genes such as p27 and p57.
All mice were housed in an air-conditioned room under adequate temperature (20.7°C-22.8°C) and humidity (41.6%-59.9%) control with a 12-hour dark/light cycle and free access to water and food (Lab diet 5002, LabDiet, Richmond, IN). All animal experiments were performed after receiving approval from the Institutional Animal Care and Use Committee (IACUC) of Clinical Research Institute at Seoul National University Hospital (SNU-070509-4), and the National Research Council (NRC) guidelines for the care and use of laboratory animals were followed (revised in 1996).
To produce miR-222/221 transgenic mice, miR-222/221 was first amplified using C57/BL6J mice. It was cloned into pGEM-T easy vector and subsequently transferred into the pcDNA3 vector using the BamHI and XhoI sites. The nucleotide sequence of the plasmid was confirmed by sequencing. The plasmid was then injected into fertilized eggs. Quantitative polymerase chain reaction screening was performed to verify that the transgene was transferred to the mice. Genomic DNA was extracted from the tail of each mouse. The presence of the miRNA-222/221 transgene was assessed by using LaboPass Tissue Mini DNA Purification Kit (Cosmo Genetech, Seoul, Korea) according to the manufacturer's protocol. The primer sequences are listed in Supplementary Table 1.
MNU (Sigma Chemical Co., St. Louis, MO) was dissolved in distilled water at a concentration of 240 ppm and added to drinking water in light-shielded bottles. It was freshly prepared thrice per week. At 6 weeks of age, mice from each group were given drinking water containing MNU on alternating weeks for a total of 10 weeks of exposure as previously described [16]. At the end of the treatment, the drinking water was replaced by autoclaved distilled water.
A total of 53 wild-type mice and 65 transgenic mice were used. At 6 weeks of age, mice from each group were treated with MNU five times during alternate weeks. The animals were carefully autopsied at 36 weeks of age, under anesthesia. At the end of the experiment, all mice were fasted for 24 hours, but had free access to water. Prior to euthanasia, blood samples were collected from the vena cava for blood chemistry tests. The stomachs and other organs were carefully examined macroscopically. After pathologic observation of the stomach and intestine, immunohistochemistry and validation studies were performed (Fig. 1).
All mice underwent a thorough postmortem examination at the time of euthanasia. The stomach was tied with silk thread at the esophagus and duodenum and then removed. Thereafter, phosphate buffered saline was injected into the stomach and 10 minutes later, the stomach was excised along the greater curvature with scissors and fixed with a pin. Half of the excised stomach was frozen in liquid nitrogen for molecular biology experiments and the remaining stomach was fixed for 24 hours, processed by standard methods, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin (H&E). The intestine was removed from the abdomen and tied with silk thread at the duodenum and the edge of the rectum. It was then injected with 10% neutral buffered formaldehyde solution. Twenty-four hours later, the knots were untied, the intestine was excised longitudinally with scissors and the contents were carefully removed. Next, the segment was rolled up longitudinally, with the mucosa outwards, using forceps [13]. Finally, the intestine was placed in a fixative for paraffin-wax embedding and sectioned. The 5-μm-thick sections were stained with H&E. Formalin-fixed, paraffin-embedded, 4-μm sections were dewaxed in xylene, rehydrated through graded alcohol, and placed in an endogenous peroxide block for 15 minutes. Sections were washed in water, antigen retrieved, and then placed in citrate buffer. Nonreactive staining was blocked by treating sections with 1% horse serum in Tris-buffered saline (pH 6.0) for 3 minutes.
Anti-CD31 (1:80, goat polyclonal, Santa Cruz Biotechnology, Santa Cruz, CA), anti–Ki-67 (1:200, rat monoclonal, DAKO, Glostrup, Denmark), anti-cleaved caspase-3 (1:100, rabbit monoclonal, Cell Signaling, Danvers, MA), anti-p27 (1:500, rabbit polyclonal, Santa Cruz Biotechnology), and anti-p57 (1:300, rabbit monoclonal, Epitomics, Burlingame, CA) were then applied. Antibody binding was detected using an avidin-biotin-peroxidase complex (Universal Elite ABC Kit, Vectastain, Burlingame, CA) for 10 minutes. Diaminobenzidine tetrahydrochloride solution (Kit HK153-5K, Biogenex, San Ramon, CA) was then used as the chromogen.
Total RNA was extracted from mouse tissues by using TRI-zol reagent (Invitrogen, Carlsbad, CA) and 30 μg was loaded on a 12.50% urea-polyacrylamide gel. The resolved RNA was transferred to a Zeta-Probe GT blotting membrane (Bio-Rad Laboratories, Hercules, CA) by capillary method and fixed by ultraviolet crosslinking. Oligonucleotides complementary to each miRNA were end-labeled at the 5′-end with 32P-γ-ATP using T4 polynucleotide kinase (Takara, Tokyo, Japan) and used as probes. Prehybridization and hybridization were carried out using ExpressHyb Hybridization Solution (Takara) according to the manufacturer's instructions. The sequences of the antisense oligos are complimentary to mature miR-222 and miR-221. The sequences of the probes are shown in Supplementary Table 1. Membranes were prehybridized for 30 minutes at 37°C using hybridization solution in hybridization tubes. Subsequently, the probes were added into the tubes and incubated for 1 hour at 37°C. After hybridization, the membranes were washed twice for 30 minutes using buffer 1 (0.05% sodium dodecyl sulphate [SDS], 2× sodium saline citrate [SSC]) at room temperature and then washed twice for 15 minutes using buffer 2 (10% SDS, 20× SSC) at room temperature. BAS 2500 Image Analysis System and Multi Gauge V3.0 software (Fujifilm, Tokyo, Japan) were used for quantitation. Ethidium bromide staining of rRNA was used for RNA loading control.
Ten human GC cell lines, SNU-638, and AGS were obtained from the Korean cell line bank. RPMI 1640 (Welgene, Daegu, Korea) supplemented with 10% fetal bovine serum (Thermo Scientific, Waltham, MA) and 1% penicillin and streptomycin (Life Technologies, Carlsbad, CA) was used for cell culture. The cells were maintained at 37°C under an atmosphere of 5% CO2-95% air.
Transfection to AGS cells was performed one day after seeding using Lipofectamine 2000 (Invitrogen).
One microgram of total RNA was used for single-stranded cDNA using the SuperScript First Strand cDNA synthesis (Life Technologies). RNA quantification was conducted by real-time polymerase chain reaction (PCR) using the comparative Ct method with the Bio-Rad iCycler Real-Time PCR system. Real-time PCR was performed in a 20 μL final volume containing 10 μL of SYBR 2× master mix (Thermo Fisher Scientific), 10 pmol of each primer in a volume of 1 μL, and 1 μL of cDNA. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primer sets were used for normalization. Each sample was run in triplicate (Bio-Rad Laboratories). Primers for the amplification of mature miR-221 and miR-222 are presented in Supplementary Table 1.
Statistical analyses were performed using SPSS ver. 13.0 (SPSS Inc., Chicago, IL) and graphs were generated using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA). Chi-square was used to measure the carcinogenesis association between the miR-222/221 transgenic and wild-type mice. p-values less than 0.05 are considered statistically significant.
The expression level of pre-miRNA-222/221 and mature miR-222/221 was increased in the stable cell line transfected with miR-222/221 as compared to the cell line transfected with pcDNA3 null vector (Fig. 2A). To examine whether off-springs contained the injected foreign DNA, germ-line transmission was tested by PCR using the CMV promoter primer and miR-222 primer at 3 weeks of age, all mice were screened by PCR (Fig. 2B). At 10 weeks of age, the expression levels of pre-miRNA-222/221 and mature miR-222/221 were compared between wild-type and transgenic mice in nine organs (brain, thymus, heart, lung, stomach, liver, intestine, spleen, and kidney) by northern blotting (Fig. 2C). miR-222/221 was expressed in transgenic mice, but not in wild-type mice. Compared to wild-type mice, transgenic mice strongly overexpressed both pre-miRNA-222/221 and mature miR-222/221 in the stomach, brain, lung, and kidney. The organ weight and blood chemistry were investigated in 56-week-old mice to compare the phenotype of transgenic and wild-type mice. There was no difference in organ weight between the two groups. When we compared body weights between male mice, transgenic mice were about 20% heavier than wild-type mice. The white blood cell (WBC) count was 1.8-fold lowerin transgenic mice in blood chemistry tests (Table 1).
MNU treated mice were euthanized at 36 weeks of age. Transgenic and wild-type mice were randomly selected and the expression levels of miR-222 and miR-221 were confirmed by northern blot and real-time PCR (Supplementary Fig. S1). Approximately 85% of transgenic mice survived until 36 weeks of age (Table 2). At the end of the experiment, all surviving mice were euthanized, and their stomachs and other organs were carefully examined. From the pathological findings, after treatment with the carcinogen MNU, carcinoma and other lesions, including hyperplasia, microcarcinoma in adenoma, angiosarcoma, microscopic dysplasia, adenoma in pylorus region, and squamous papilloma in the fundus region, were detected in the mice stomachs. However, only flat serrated adenoma was observed from the intestines of three mice. From these results, adenoma and hyperplasia, the two most frequently observed pre-cancerous lesions, were selected and included for MNU-induced multigastric carcinogenesis steps. Representative histological images of normal, pre-cancerous lesions and carcinoma are shown in Fig. 3. Hyperplasia was observed in 3.77% of the wild-type mice and in 18.46% of the transgenic mice (p=0.020). Adenoma was observed in 20.75% of the wild-type mice and 26.15% of the transgenic mice (p=0.522). Carcinoma was observed in 32.08% of the wild-type mice and 41.54% of the transgenic mice (p=0.341). The frequency of hyperplasia, adenoma, and carcinoma was higher in transgenic mice, but the difference was statistically significant only in hyperplasia (Table 3). When hyperplasia and adenoma were combined as a pre-cancerous lesion, it was observed in 44.62% of transgenic mice and 24.53% of wild-type mice; this difference was statistically significant (p=0.033).
To test the correlation between cancer and miR-222/221 in our in vivo model, cancer related markers were selected for immunohistochemistry. CD31 was used as a marker of angiogenesis, Ki-67 was used to assess cell proliferation, and cleaved caspase 3 was used as an apoptosis marker. Moreover, two antibodies against proteins encoded by two miR-222/221 target genes, p27 and p57, were used to compare p27 and p57 protein expression in the stomachs of transgenic and wild-type mice by immunohistochemistry. Based on our hypothesis, in transgenic mice overexpressing miR-222/221, the expression of p27 and p57 should be repressed as compared to that in wild-type mice. However, there was no significant difference in term of p27 and p57 expression between the transgenic and wild-type mice. Compared to wild-type mice, in transgenic mice, CD31 (angiogenesis marker) and Ki-67 (proliferation marker) were highly expressed, suggesting that cancer-related biological processes, angiogenesis and proliferation, were activated. However, the expression of cleaved caspase 3 (apoptosis marker), p27, and p57 was not significantly different between the two groups (Fig. 4A). Additionally, p27 expression was similar in the stomach of untreated wild-type and transgenic mice (Fig. 4B).
Despite the significant number of patients with gastric cancer, surgery is often the only treatment option because of the many side effects of chemotherapy and radiation therapy [8,17]. Surgery is also limited to local treatment. For this reason, miRNA has gained much attention due to its assumed role in carcinogenesis. However, the molecular pathology of gastric cancer remains poorly understood even though gastric cancer is a major health burden throughout the world. Therefore, the study of miRNAs implicated in gastric cancer formation is urgent and may provide opportunities to develop new means for diagnosis and treatment of gastric cancer. In addition, the accurate function of miRNAs in vivo still need to be further investigated. Many groups attempted to generate miRNA knockout or transgenic mice to validate the specificity and efficiency of microRNA in vivo animal models.
Regarding the phenotype of the miR-222/221 transgenic mice, the body weight of male transgenic mice was about 20% higher than that of the control group whereas no significant difference was observed in term of body weight between transgenic and wild-type female mice. The result of blood testing indicated unique differences in WBC and monocytes. Levels of WBC and monocytes were significantly lower by 1.8-fold and 1.3-fold, respectively, in transgenic mice than in wild-type mice. WBC and monocytes are related to immune response and, therefore, aberrantly low levels of WBC and monocytes result in a decreased ability to fight virus or other pathogens. This shortage of immune response may contribute to the enhanced gastric cancer development in miR-222/221 transgenic mice. After euthanasia, the Swiss roll technique was performed on all mice to detect abnormal intestinal lesions. No carcinoma or cancer-related lesions were detected in the intestine, but flat serrated adenoma was observed in three transgenic mice.
When the phenotype of transgenic mice was examined at 56 weeks of age, no aberrant lesions were observed macroscopically and during histological examination. This result suggested that miR-222/221 overexpression in these transgenic mice was not sufficient to cause gastric carcinogenesis by itself. Therefore, MNU, which is a well-known rodent carcinogenic, was administered to both transgenic and wild-type mice to promote gastric carcinogenesis using a previously described protocol [14-16,18].
The MNU-induced mouse models for gastric carcinogenesis have been widely used to study carcinogenesis of the stomach [14-16,18]. These animal models have been used not only to investigate the pathogenesis of gastric carcinogenesis, but also to identify tumor promoters and chemopreventive agents [16,19]. To determine the accurate time at which carcinogenesis and cancer-related lesions could be detected, five MNU-treated mice from each group were euthanized at 27 weeks of age, but no significant histological difference between transgenic and wild-type mice was observed. This can be explained by the fact that mice are known to be relatively resistant to MNU. Therefore, a longer period is required to induce gastric carcinoma in C57BL/6J mice [15,19-21]. When the MNU-treated mice were euthanized at 36 weeks of age, MNU-induced gastric carcinogenesis steps were detected.
After euthanasia at 36 weeks of age, pathological examination was performed to evaluate whether the lesions observed were related to cancer. The alkylating agent MNU induces the development of different patterns of cancer-related lesions. For instance, hyperplasia is frequently observed when using MNU, as previously reported by many groups [14,15,19]. The reason why hyperplasia is often detected in MNU-induced gastric carcinogenesis models is not fully understood, but it is commonly found in other carcinogen-induced gastric carcinogenesis models. The result was also similar to macroscopic findings. The whole stomach of the mice was thickened and stiff with pre-cancerous lesions.
Recent studies revealed that miR-222/221 is overexpressed in several types of cancer and has oncogenic effects [22-24]. In most cases, in vitro studies have been reported, but one group recently developed an in vivo model of liver cancer. The transgenic model, in which miR-221 is expressed specifically in the liver, was developed and characterized by the appearance of spontaneous liver tumors in a fraction of male mice and a strong acceleration of tumor development in 100% of mice treated with diethylnitrosamine [25]. However, in our study, carcinoma occurrence was not statistically significant, while frequently observed pre-cancerous lesions were statistically significant. In our transgenic model, the occurrence of carcinoma could not be dramatically induced. This can be explained by the fact that we did not use a promoter specific for the stomach and could not transcriptionally control the expression of miR-222/221 when the transgenic mouse model was developed. In the case of the miR-221 liver cancer transgenic mouse, a liver-specific promoter was used and the overexpression of miR-221 was induced. Our transgenic mice expressed miR-222/221 in all organs. Thus, showing a strong effect of miR-222/221 in the stomach was expected to be difficult.
Ultimately, even though assessing the expression of p27 was difficult because of the changes (mRNA, protein, and immunohistochemistry) induced by MNU, we demonstrated that the gastric carcinogenic effect of miR-222/221 with the help of MNU was more frequently detected in transgenic mice than in wild-type mice, although it was a mild effect.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (http://www.e-crt.org).
ACKNOWLEDGMENTS
This study was supported by grants from the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI13C2148).
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–54.

2. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008; 9:102–14.

3. Meng F, Hackenberg M, Li Z, Yan J, Chen T. Discovery of novel microRNAs in rat kidney using next generation sequencing and microarray validation. PLoS One. 2012; 7:e34394.

4. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105.

5. Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005; 132:4653–62.

6. Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008; 27:5651–61.

7. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55:74–108.

8. Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008; 9:279–87.

9. Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010; 70:2339–49.
10. Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009; 24:652–7.

11. Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008; 13:272–86.
12. Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008; 123:372–9.

13. Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009; 37:1672–81.

14. Yoshizawa N, Yamaguchi H, Yamamoto M, Shimizu N, Furihata C, Tatematsu M, et al. Gastric carcinogenesis by N-Methyl-N-nitrosourea is enhanced in db/db diabetic mice. Cancer Sci. 2009; 100:1180–5.
15. Tomita H, Yamada Y, Oyama T, Hata K, Hirose Y, Hara A, et al. Development of gastric tumors in Apc (Min/+) mice by the activation of the beta-catenin/Tcf signaling pathway. Cancer Res. 2007; 67:4079–87.
16. Yamachika T, Nakanishi H, Inada K, Tsukamoto T, Shimizu N, Kobayashi K, et al. N-methyl-N-nitrosourea concentration-dependent, rather than total intake-dependent, induction of adenocarcinomas in the glandular stomach of BALB/c mice. Jpn J Cancer Res. 1998; 89:385–91.

17. Jiang J, Xu N, Wu C, Deng H, Lu M, Li M, et al. Treatment of advanced gastric cancer by chemotherapy combined with autologous cytokine-induced killer cells. Anticancer Res. 2006; 26:2237–42.
18. Hahm KB, Song YJ, Oh TY, Lee JS, Surh YJ, Kim YB, et al. Chemoprevention of Helicobacter pylori-associated gastric carcinogenesis in a mouse model: is it possible? J Biochem Mol Biol. 2003; 36:82–94.

19. Lu J, Imamura K, Nomura S, Mafune K, Nakajima A, Kadowaki T, et al. Chemopreventive effect of peroxisome proliferator-activated receptor gamma on gastric carcinogenesis in mice. Cancer Res. 2005; 65:4769–74.
20. Nam KT, Hahm KB, Oh SY, Yeo M, Han SU, Ahn B, et al. The selective cyclooxygenase-2 inhibitor nimesulide prevents Helicobacter pylori-associated gastric cancer development in a mouse model. Clin Cancer Res. 2004; 10:8105–13.

21. Han SU, Kim YB, Joo HJ, Hahm KB, Lee WH, Cho YK, et al. Helicobacter pylori infection promotes gastric carcinogenesis in a mice model. J Gastroenterol Hepatol. 2002; 17:253–61.

22. le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007; 26:3699–708.

23. Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007; 282:23716–24.

Fig. 1.
Schematic diagram of the study design indicating that mice were observed for 36 weeks, followed by immunohistochemical analysis. MNU, N-nitroso–N–methylurea.
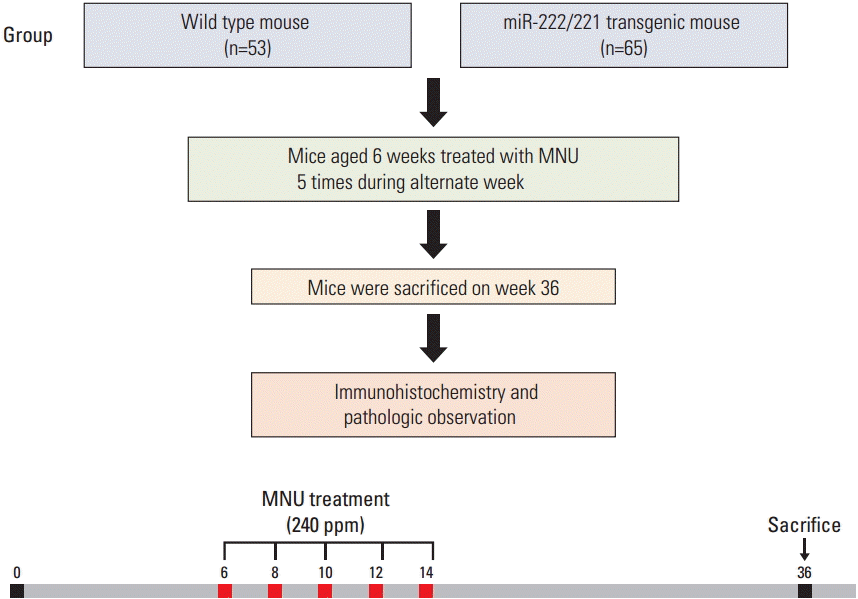
Fig. 2.
Phenotype of miR-222/221 transgenic (TG) mice and selection. (A) miR-222/221 expression was confirmed in the stable cell line by northern blot analysis. Pre-miRNA-222/221 and mature miRNA-222/221 levels were increased in the stable cell line transfected with miR-222/221. (B) miR-222/221 expression was assessed by polymerase chain reaction analysis with the CMV promoter and miR-222 primers in offsprings to select TG or wild-type (WT) mice. Mouse 1, 2, and 4 were generated from a TG mouse. (C) miR-222/221 expression level in various organs from miR-222/221 TG and WT mice.
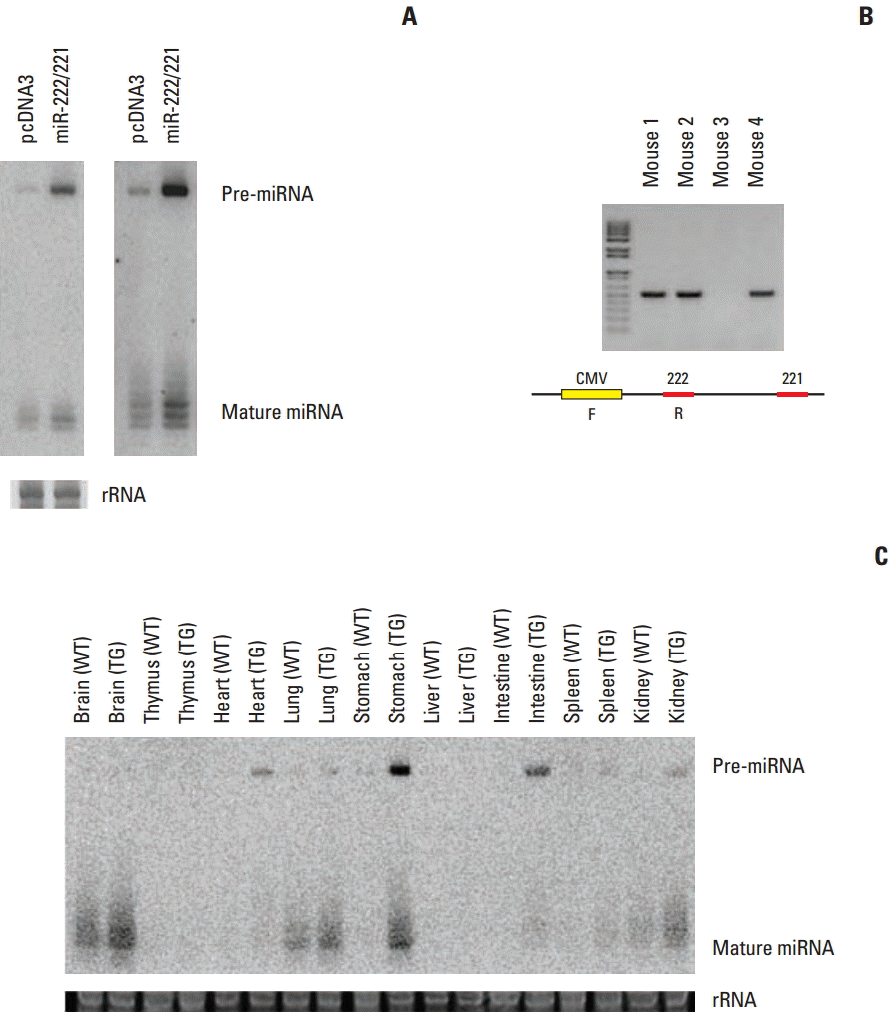
Fig. 3.
Representative histological images of normal, pre-cancerous lesions, and carcinoma in transgenic and wild-type mice (A, C, D, H&E staining, ×40; B, H&E staining, ×100).
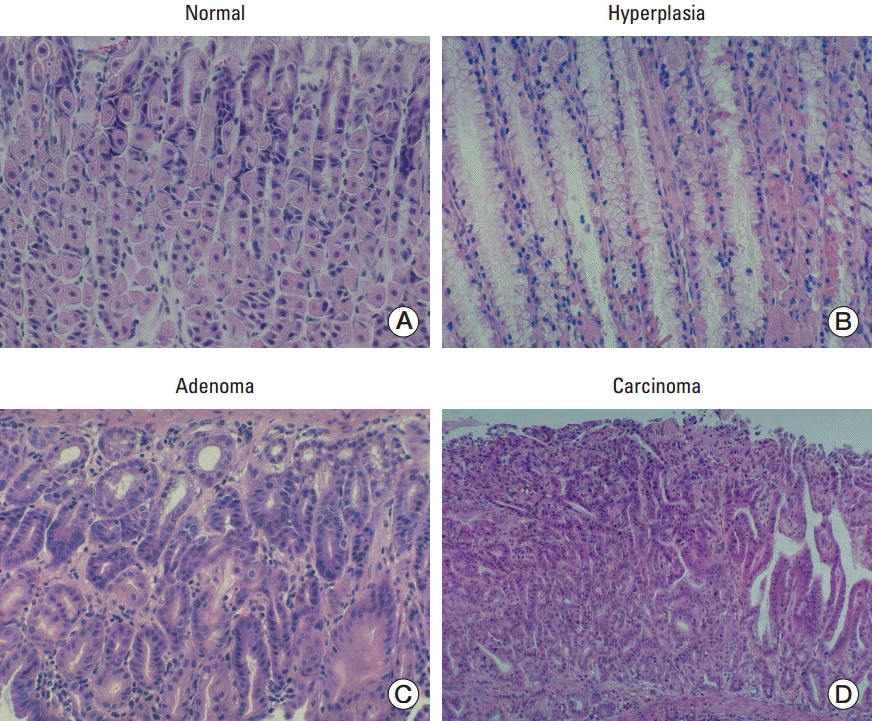
Fig. 4.
Cancer-related markers analyzed by immunohistochemistry. (A) Histological analysis of the stomachs of MNU-treated mice. CD31 and Ki-67 expression increased in transgenic (TG) mice compared to that in wild-type (WT) mice (×100). No significant difference was observed in caspase 3, p57, and p27 expression. (B) Histological analysis of the stomachs of MNU-untreated mice. p27 expression was similar in the stomachs of WT and TG mice.
Table 1.
Organ weight and blood chemistry test of miR-222/221 transgenic mice (56-week-old)
Table 2.
Mortality within 30 weeks and MNU intake of mice according to group
| Group | Total No. of mice | No. of mice finally evaluated | Dead No. of mice (%) |
|---|---|---|---|
| WT mouse | 59 | 53 | 6 (10.16) |
| TG mouse | 76 | 65 | 11 (14.47) |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download