This article has been
cited by other articles in ScienceCentral.
Abstract
Background
Immunohistochemistry (IHC) has played an essential role in the diagnosis of hematolymphoid neoplasms. However, IHC interpretations can be challenging in daily practice, and exponentially expanding volumes of IHC data are making the task increasingly difficult. We therefore developed a machine-learning expert-supporting system for diagnosing lymphoid neoplasms.
Methods
A probabilistic decision-tree algorithm based on the Bayesian theorem was used to develop mobile application software for iOS and Android platforms. We tested the software with real data from 602 training and 392 validation cases of lymphoid neoplasms and compared the precision hit rates between the training and validation datasets.
Results
IHC expression data for 150 lymphoid neoplasms and 584 antibodies was gathered. The precision hit rates of 94.7% in the training data and 95.7% in the validation data for lymphomas were not statistically significant. Results in most B-cell lymphomas were excellent, and generally equivalent performance was seen in T-cell lymphomas. The primary reasons for lack of precision were atypical IHC profiles for certain cases (e.g., CD15-negative Hodgkin lymphoma), a lack of disease-specific markers, and overlapping IHC profiles of similar diseases.
Conclusions
Application of the machine-learning algorithm to diagnosis precision produced acceptable hit rates in training and validation datasets. Because of the lack of origin- or disease-specific markers in differential diagnosis, contextual information such as clinical and histological features should be taken into account to make proper use of this system in the pathologic decision-making process.
Keywords: Database, Expert-supporting system, Machine learning, Immunohistochemistry, Probabilistic decision tree
Immunohistochemical (IHC) staining is a valuable and unique tissue-staining method for pathologic diagnosis of hematolymphoid neoplasms using an antigen-antibody reaction [
1-
6]. It was developed as an indirect immunofluorescence technique in 1941 by Albert Coons at Harvard University [
1,
7]. As the IHC technique evolved to use paraffin-embedded tissues and enzymatic markers, it has become an essential and routine tool in pathologic diagnoses, notably in hematopathology [
2-
6]. Hematolymphoid neoplasms are categorized as B-cell, T-cell, NK/T-cell, and histiocytic neoplasms according to the IHC profiles of CD3 (T-cell marker), CD20 (B-cell marker), CD56 (NK-cell marker), CD68 (histiocytic marker), and other markers related to the development of hematolymphoid cells. Once the morphologic features that distinguish among the possible groups are recognized in differential diagnosis, relevant IHC panels can be chosen for subtype determination (
Supplementary Fig. S1). This process requires the pathologists’ intuition and comprehensive integration of both the clinicopathologic findings and IHC results because hematolymphoid neoplasms share many cytomorphologic and clinicopathologic features across different diseases. The accurate subtyping of lymphomas is therefore highly dependent on the appropriate choice of IHC panels and the interpretation of IHC results [
1-
3,
8].
However, increasing knowledge of IHC positivity in each tumor can produce conflicting interpretations in daily practice, especially in some more complex cases [
9]. The pathologic analysis of IHC results depends largely on the expertise of pathologists, who can be easily biased by their experiences [
2,
4,
6]. New antibodies and IHC data from various tumors are introduced annually, and more than 100,000 studies using IHC have been published since 2000, making it difficult to memorize the newly developed antibodies and recognize the expression characteristics of tumors just in the human brain [
10-
14]. In addition, recent advances in digital pathology require an appropriate reference database of ancillary tests to integrate medical knowledge and individual medical problems [
15].
Attempts have been made to address this problem by adopting an algorithmic approach or using standardized IHC panels for specific differential diagnosis [
9,
14,
16]. However, the clinical situation of each case is unique, and generalized application of a particular IHC panel or specific algorithm can be time- and laborconsuming.
We therefore developed an expert-supporting system using software based on a machine-learning algorithm and an IHC database that supports pathological decision-making and differential diagnosis. We developed the software as a mobile application for iOS and Android devices for practical utility.
MATERIALS AND METHODS
Development of a machine-learning algorithm using a probabilistic decision tree
According to Bayes’ theorem, post-event probability can be calculated when pre-event probability is given. Bayes’ theorem is stated mathematically as
where A and B are events and P(B)≠016 and P(A|B), and P(B|A) are the respective conditional probability that the likelihood of event A occurring given that B is true and vice versa. P(A) and P(B) are the probabilities of observing A and B independently of each other [
16,
17].
A probabilistic decision tree is a predictive modeling approach in statistics and data mining. It is often used for machine-learning algorithms, especially when test node results are binary (
Fig. 1). We adopted such a tree for our machine-learning algorithm because IHC results are binary, and the probability can be expressed as a database.
To apply the probabilistic decision-tree algorithm, we required a database of a 2×2 table with tests, diseases, and the probability of positivity of each test for each disease (
Fig. 2). Test results were binary, and the probability of positivity was the number of positive cases among all cases of the disease. Once test results were available, we calculated the probability of each disease by multiplying prior probability by the probability that each test was positive or negative to determine the most probably illness by comparing post-probabilities.
IHC database build
We assembled a database of IHC expression profiles of the lymphoid neoplasms using five primary textbooks and other publications, including the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues (IARC, Lyon, France) and major IHC textbooks (
Supplementary Table S1) [
4,
5,
18-
21]. More than 200 lymphoid neoplasms and tumor names were documented according to the WHO classification. Tumors without IHC profile data or no diagnostic use were excluded. Subtypes of certain tumors were documented separately from the primary type if there was a difference in IHC profiles.
The IHC positivity for each tumor was drawn from textbook descriptions; if there was exact numerical value was replaced by arbitrary expressions such as “always positive,” “often positive,” or “rarely/occasionally positive,” positivity was documented as follows: always, 95%; often, 75%; in about a half of cases, 50%; seldom, 30%; rarely/occasionally, 10%; never, 0%. If the positivity differed between textbooks, the average value was used. An example of the IHC database appears in
Supplementary Fig. S2.
Approximately 600 IHC antibody names were documented using the textbooks, and their synonyms were recorded and revised with online references (
Supplementary Table S2).
Development of a mobile application
The reactive native (network-free) mobile application “ImmunoGenius” was developed using NoSQL for iOS and Android (
Fig. 3), which can be used on iOS and Android devices. It was designed to search and select diseases and generate a 2×2 table with the disease name in the left column and IHC antibody names on the first row. Representative IHC profiles appear in the corresponding cells as ++ for 75%–100% positivity (positive cases per all tumors), + for 50%–74%, +/– for 30%–49%, –/+ for 10%–29%, and – for 0%–9% with graded shades (
Fig. 3,
Supplementary Fig. S3,
Supplementary Video 1). Users can compare IHC profiles between the selected diseases and add or remove rows (diseases) or columns (IHC antibodies) to customize the table. Additional IHCs can be added using the buttons on the right side. Once the user inputs the IHC results for their case, the 10 most probable diagnoses as calculated by the diagnosis precision algorithm appear below, along with estimated probability in percentage (red numbers) (
Supplementary Fig. S3,
Supplementary Video 1,
2). For predictive diagnosis of lymphomas, prior probability was set according to epidemiologic data for Korea in 2010 (but can be changed to other epidemiologic groups later).
Diagnosis precision algorithm validation using patient data
To validate the hit rate of the diagnosis precision algorithm, the IHC profile data and diagnoses originally made by pathologists were compared with the top 10 predictive diagnoses produced by the algorithm. Approximately 1,000 cases of the lymphoma-patient IHC profile data were obtained from the archives of two independent university hospitals: Yeouido, and Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, from 2010 to 2017. Approximately 80 percent of the lymphoma cases at Seoul St. Mary’s Hospital were referred from various institutes in Korea. Any patient data related to identification except the original diagnosis and the IHC results were blinded before data processing. The retrieved data were divided 6:4 for training and validation. Cases with an inconclusive diagnosis or inadequate IHC profile (fewer than three antibodies, inconclusive results, absence of markers for tumor origins, but only prognostic or therapeutic markers such as epidermal growth factor receptor or p53) were excluded. An example of a retrieved patient IHC profile dataset appeared in
Supplementary Fig. S4. The diagnosis precision hit rate was determined by the inclusion of the original diagnosis in the top 10 predictive diagnoses drawn by the algorithm. It was considered inclusive if there was no significant difference in the IHC profile between the original and predictive diagnosis, and the only difference was in location if two diagnoses shared the same origin of cells (e.g., nodal marginal zone lymphoma vs. extranodal marginal zone lymphoma). Validation of the algorithm was carried out by comparing the hit rate of the training and validation data in lymphomas. If no statistically significant difference was found between the training and validation dataset, the algorithm was considered validated.
Statistical analysis
Time and computational complexity were evaluated by testing the mobile application. The hit rate between original and predictive diagnoses was compared by chi-square tests. Statistical analysis was performed using Web-R (“
http://web-r.org”), a webbased statistical analysis program.
RESULTS
IHC database build and recruitment of training and validation datasets
A total of 150 hematolymphoid neoplasms and 584 IHC antibodies and their IHC profiles were documented. The obtained training and validation data of lymphoma amounted to 639 and 392 cases, respectively. In the lymphoma cases, an average of 8.5 IHC antibodies (range, 1 to 18) were used for diagnosis, and 40 types of lymphomas were included. Two cases were excluded because of inconclusive diagnosis, and 35 cases with fewer than 3 IHC tested antibodies were omitted. As a result, 602 cases of lymphomas were used for training. The original diagnoses of the training data cases are provided in
Table 1. Diffuse large B-cell lymphoma, not otherwise specified (DLBCL, NOS) was the most common, with 216 cases (34.3%), and the second most common was extranodal marginal zone lymphoma of mucosaassociated lymphoid tissue (MALT), with 78 cases (13.0%). The original diagnoses of the validation cases are also provided in
Table 1. The most and the second most common type were the same as DLBCL, NOS, and MALT lymphoma, with similar percentages (145, 37.0% and 74, 18.9%, respectively).
Training data
The hit rate for training data of the predictive diagnosis (top 10) was 94.7% (
Table 2). Detailed results of discordant cases between the original and predictive diagnoses are supplied in
Table 2. In B-cell lymphomas, the hit rate of the predictive diagnosis was relatively high, particularly in DLBCL, follicular lymphoma, chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and MALT lymphoma with zero error rates. The diagnoses showed generally good performance in most B-cell lymphomas, with the exception of plasmablastic lymphoma (three errors out of three cases, 100%) and one mantle cell lymphoma (MCL). In T-cell lymphomas, the algorithm achieved a performance that is generally equivalent to that in B-cell lymphomas except for T lymphoblastic leukemia/lymphoma (no error in 17 cases) and extranodal NK/T-cell lymphoma, nasal type (no error in 25 cases). In enteropathy-associated T-cell lymphoma, peripheral T-cell lymphoma, NOS, anaplastic large cell lymphoma (ALCL), anaplastic lymphoma kinase (ALK)–negative, and angioimmunoblastic T-cell lymphoma, the error rates were 50.0%, 34.7%, 43.8%, and 33.3%, respectively. In Hodgkin lymphomas, the error rates were 12.5% in classical Hodgkin lymphoma, NOS, and 14.3% in nodular sclerosis subtype.
Validation data
In the validation data, the hit rate of the predictive diagnosis (top 10) was 95.7% (
Table 2). Detailed results of the discordant cases for the original and predictive diagnoses are provided in
Table 2. In B-cell lymphomas, the hit rate of predictive diagnosis was relatively high for DLBCL, follicular lymphoma, CLL/SLL, MALT lymphoma, and MCL with zero error rates. Generally good performance was seen in most B-cell lymphomas, with the exception of primary cutaneous follicle center lymphoma (1 error out of 2 cases, 50.0%). In T-cell lymphomas, performance was generally equivalent to that in B-cell lymphomas with the exception of T lymphoblastic leukemia/lymphoma (no errors in 7 cases), extranodal NK/T-cell lymphoma, and nasal type (no errors in 15 cases). In enteropathy-associated T-cell lymphoma, primary cutaneous CD8-positive, aggressive epidermotropic cytotoxic T-cell lymphoma, peripheral T-cell lymphoma, NOS, ALCL, ALK-negative, and angioimmunoblastic T-cell lymphoma, the error rates were 50.0%, 50.0%, 33.3%, 42.8%, and 33.3%, respectively. In Hodgkin lymphomas, the error rates were 50.0% in nodular lymphocyte-predominant Hodgkin lymphoma, 18.2% in classical Hodgkin lymphoma, NOS, 20.0% in nodular sclerosis subtype, and 33.3% in lymphocyte-depleted subtype.
Precision error rates between training and validation dataset
The error rates of the predictive diagnosis were 5.3% in training data and 4.3% in validation data. The error rates of both groups were not significantly different (p=0.543) (
Table 3). The overall hit rate was 95.0% in lymphomas (
Table 3).
DISCUSSION
We verified that it is possible to calculate the probability of a specific disease for a particular case, especially lymphomas, using IHC results, a probabilistic decision tree, and a mobile application. The diagnosis precision drawn by the probabilistic decision-tree algorithm achieved a hit rate of 95.0% for lymphomas. The hit rates between training and validation dataset did not differ significantly in lymphomas (94.7% vs. 95.7%, p=0.543).
The hit rate of the diagnosis precision algorithm was relatively high in most B-cell lymphomas, including DLBCL, follicular lymphoma, CLL/SLL, MALT lymphoma, and Burkitt lymphoma with zero errors, which represents the majority of all lymphoma cases (approximately two-thirds). One case of MCL showed an incorrect predictive diagnosis that was an atypical case of a cyclinD1-negative MCL with a CCND1/IGH translocation proven by fluorescence in situ hybridization (IHC results; CD20+, Bcl-2+, CD3–, CD10–, Bcl-6–, CD23–, MUM1–, p53–). The IHC for CD5 was not available, and this atypical IHC profile appeared to explain the incorrect precision. Another case of cyclinD1-negative MCL occurred in the validation data set. Although the cyclinD1 was negative, CD5 was positive, and the predictive diagnosis included MCL. In plasmablastic lymphomas, the predictive diagnosis was incorrect in all three cases. Plasmablastic lymphoma shares an IHC profile with plasma cell neoplasms, large B-cell lymphomas, and MALT lymphoma, in that CD38, CD138, CD79a, are positive and CD30 is positive but CD20 is often negative [
5,
18,
19,
20]. All three recruited cases of plasmablastic lymphoma showed CD20 positivity and were presumed to be plasma cell neoplasms such as multiple myeloma and solitary plasmacytoma, diffuse large B-cell lymphoma, anaplastic variant (CD30 positive), and extranodal MALT lymphoma with plasmacytoid differentiation. The main reason for the error in diagnosis precision is thought to be a lack of diseasespecific markers and overlapping IHC profiles similar to those of other diseases. Likewise, in primary cutaneous follicle center lymphoma of the validation dataset, the similarity of IHC profiles to follicular lymphomas appeared to explain the lack of precision. Because the algorithm does not take into account clinicopathologic information such as tumor location, skin versus lymph node, in this case, this incoherence can be explained.
In T-cell lymphomas, by comparison, the algorithm achieved a generally equivalent performance compared with B-cell lymphomas, with the exception of T lymphoblastic leukemia/lymphoma and extranodal NK/T-cell lymphoma, and nasal type. The accurate precision in T lymphoblastic leukemia/lymphoma and extranodal NK/T-cell lymphoma in the nasal type is due to the presence of disease-specific markers such as TdT, and CD56, and Epstein-Barr virus–encoded small RNA. However, in adult T-cell leukemia/lymphoma, enteropathy-associated T-cell lymphoma, primary cutaneous CD8-positive, aggressive epidermotropic cytotoxic T-cell lymphoma, peripheral T-cell lymphoma, NOS, ALCL, ALK-negative, and angioimmunoblastic T-cell lymphoma, error rates were high as 33.3% to 100.0%. Adult T-cell leukemia/lymphoma shares an IHC profile with peripheral T-cell lymphoma, NOS, with no disease-specific markers, but distinctive clinicopathologic features [
18-
20]. Enteropathy-associated T-cell lymphomas also do not have pathognomic IHC markers but distinctive clinicopathologic findings and often share an IHC profile with peripheral T-cell lymphomas [
18-
20]. Primary cutaneous CD8-positive, aggressive epidermotropic cytotoxic T-cell lymphoma is a rare subtype of peripheral T-cell lymphoma that involves primarily skin, and the IHC profile is not specific enough to rule out other diseases by IHC alone [
4,
5,
20]. ALCL, ALK-negative is a lymphoma of anaplastic morphology with negative ALK, which can often share IHC profiles with ALCL, ALK-positive, Hodgkin lymphomas, and peripheral T-cell lymphoma, NOS [
18-
20]. In angioimmunoblastic T-cell lymphoma, programmed death-1 (PD-1) has been considered a specific marker [
22]. However, many other lymphomas often express PD-1 at varying rates, and its positivity is often interpreted based on characteristic histologic features [
4,
5,
20]. Peripheral T-cell lymphoma, NOS, is a different category of nodal and extranodal mature T-cell lymphomas that do not correspond to any explicitly defined entities by definition [
18,
19]. Therefore, its IHC profiles cover a wide variety of expressions and are often shared by other entities in T-cell lymphomas. In summary, many T-cell lymphomas often share IHC profiles and have no disease-specific IHC markers but can be differentially diagnosed based on clinicopathologic findings with or without IHC profiles.
In Hodgkin lymphomas, the error rates were 50.0% in nodular lymphocyte-predominant Hodgkin lymphoma (1 out of 2), 15.8% in classical Hodgkin lymphoma, NOS (3 out of 19), 15.4% in nodular sclerosis subtype (4 out of 26), 0% in mixed cellularity subtype (0 out of 15), and 33.3% in lymphocyte-depleted subtype (1 out of 3). Nodular lymphocyte-predominant Hodgkin lymphoma shares the IHC profile with T-cell/histiocyte-rich DLBCL and ALCL, ALK-negative, as well as clinicopathologic features. Classical Hodgkin lymphoma, including subtypes, also showed overlapping IHC profiles to peripheral T-cell lymphoma, NOS, but a differential diagnosis based only on IHC profiles is not feasible, particularly if CD15, a specific Hodgkin’s marker, is negative. Integrated and comprehensive diagnosis, including the clinicopathologic findings in addition to the possible diagnosis by IHC profiles is therefore essential.
In terms of time and user experience, it is difficult to mathematically compare the amount of time that is consumed during the process of diagnosis with or without using this application. In general, however, most pathologists gave us positive feedback about time-saving and easy-to-use user experiences.
This study demonstrates the feasibility and clinical utility of the diagnosis precision algorithm and corresponding mobile application in differential diagnosis of lymphomas using IHC profiles. The overall hit rate of this machine-learning algorithm was 95.0% in lymphomas, and the hit rates were not significantly different between training and validation data in lymphoma, which showed a relatively good generalization. Significant errors were associated with atypical IHC profiles, a lack of site- and disease-specific markers, overlapping IHC profiles between disease entities, mixed/combined tumors, etc. Although this system will help pathologists make better decisions during pathologic diagnosis by supplying comprehensive IHC information relevant to efficient and accurate differential diagnosis, integrated interpretation with contextual information such as clinical and pathological findings are recommended, and the supportive use of this application is desirable. Further studies of possible recommendations for IHC panels for specific situations involving differential diagnosis and application of artificial neural network algorithms are required to optimize the algorithm’s sensitivity to disease, organ incidence, and antibody weight.
ACKNOWLEDGMENTS
I appreciate Myungjin Choi, Dasom X Inc. for technical support in the development of mobile application and Young Dong Seo for reviewing the manuscript style.
References
1. Elias JM. Immunohistochemistry: a brief historical perspective: commentary. In : Shi SR, Gu J, Taylor CR, editors. Antigen retrieval techniques: immunohistochemistry and molecular morphology. Natick: Eaton Pub;2000. p. 7–13.
2. Buchwalow IB, Bocker W. Immunohistochemistry: basics and methods. Heidelberg: Springer;2010.
3. Matos LL, Trufelli DC, de Matos MG, da Silva Pinhal MA. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights. 2010; 5:9–20.
4. Chu P, Weiss L. Modern immunohistochemistry. 2nd ed. Cambridge: Cambridge University Press;2014.
5. Dabbs DJ. Diagnostic immunohistochemistry: theranostic and genomic applications. 4th ed. Philadelphia: Elsevier/Saunders;2014.
6. Kalyuzhny AE. Immunohistochemistry: essential elements and beyond. Cham: Springer;2016.
7. Coons AH. Labeling Techniques in the diagnosis of viral diseases. Bacteriol Rev. 1964; 28:397–9.

8. Werner B, Campos AC, Nadji M, Torres LF. Practical use of immunohistochemistry in surgical pathology. J Bras Patol Med Lab. 2005; 41:353–64.
9. DeYoung BR, Wick MR. Immunohistologic evaluation of metastatic carcinomas of unknown origin: an algorithmic approach. Semin Diagn Pathol. 2000; 17:184–93.
10. Bishop JA, Sharma R, Illei PB. Napsin A and thyroid transcription factor-1 expression in carcinomas of the lung, breast, pancreas, colon, kidney, thyroid, and malignant mesothelioma. Hum Pathol. 2010; 41:20–5.

11. Yu H, Li L, Liu D, Li WM. Expression of TTF-1, napsinA, P63, CK5/6 in lung cancer and its diagnostic values for histological classification. Sichuan Da Xue Xue Bao Yi Xue Ban. 2017; 48:336–41.
12. El-Maqsoud NM, Tawfiek ER, Abdelmeged A, Rahman MF, Moustafa AA. The diagnostic utility of the triple markers Napsin A, TTF-1, and PAX8 in differentiating between primary and metastatic lung carcinomas. Tumour Biol. 2016; 37:3123–34.

13. Gweon HM, Kim JA, Youk JH, et al. Can galectin-3 be a useful marker for conventional papillary thyroid microcarcinoma? Diagn Cytopathol. 2016; 44:103–7.

14. Kandalaft PL, Gown AM. Practical applications in immunohistochemistry: carcinomas of unknown primary site. Arch Pathol Lab Med. 2016; 140:508–23.

15. Nam S, Chong Y, Jung CK, et al. Introduction to digital pathology and computer-aided pathology. J Pathol Transl Med. 2020; 54:125–34.

16. Lin F, Prichard J. Handbook of practical immunohistochemistry: frequently asked questions. 2nd ed. New York: Springer;2015.
17. Lesaffre E, Lawson AB. Bayesian biostatistics. Chichester: John Wiley & Sons;2012.
18. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: International Agency for Research on Cancer;2008.
19. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Lyon: International Agency for Research on Cancer;2017.
20. Rekhtman N, Bishop JA. Quick reference handbook for surgical pathologists. Heidelberg: Springer;2011.
22. Dorfman DM, Brown JA, Shahsafaei A, Freeman GJ. Programmed death-1 (PD-1) is a marker of germinal center-associated T cells and angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2006; 30:802–10.

Fig. 1.
A probabilistic decision tree for a machine-learning algorithm in diagnostic tests and disease.
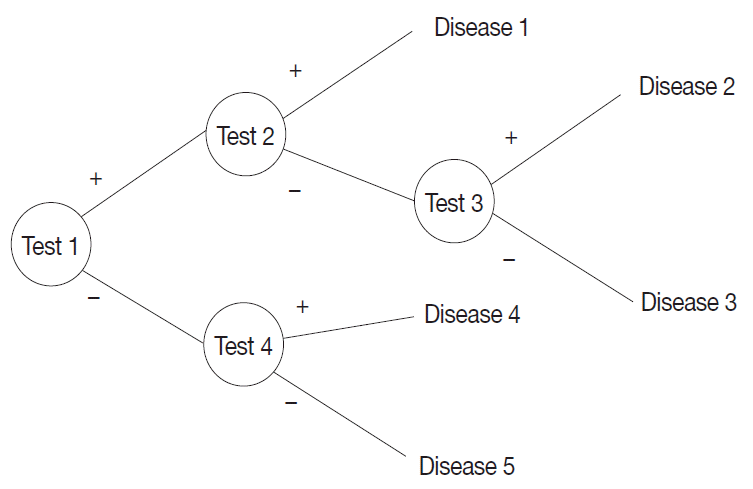
Fig. 2.
Prior and post probability based on Bayes’ theorem.
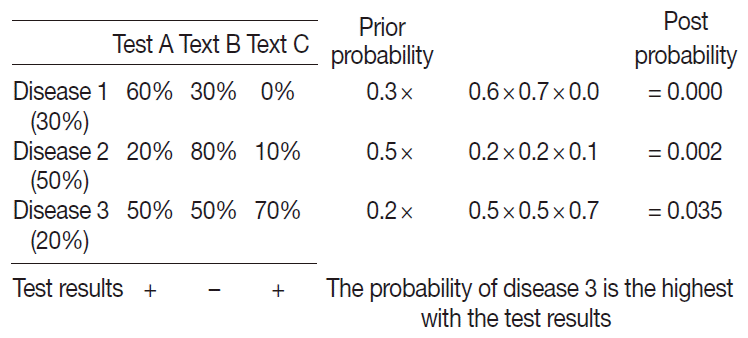
Fig. 3.
A screenshot of the mobile application “ImmunoGenius.”
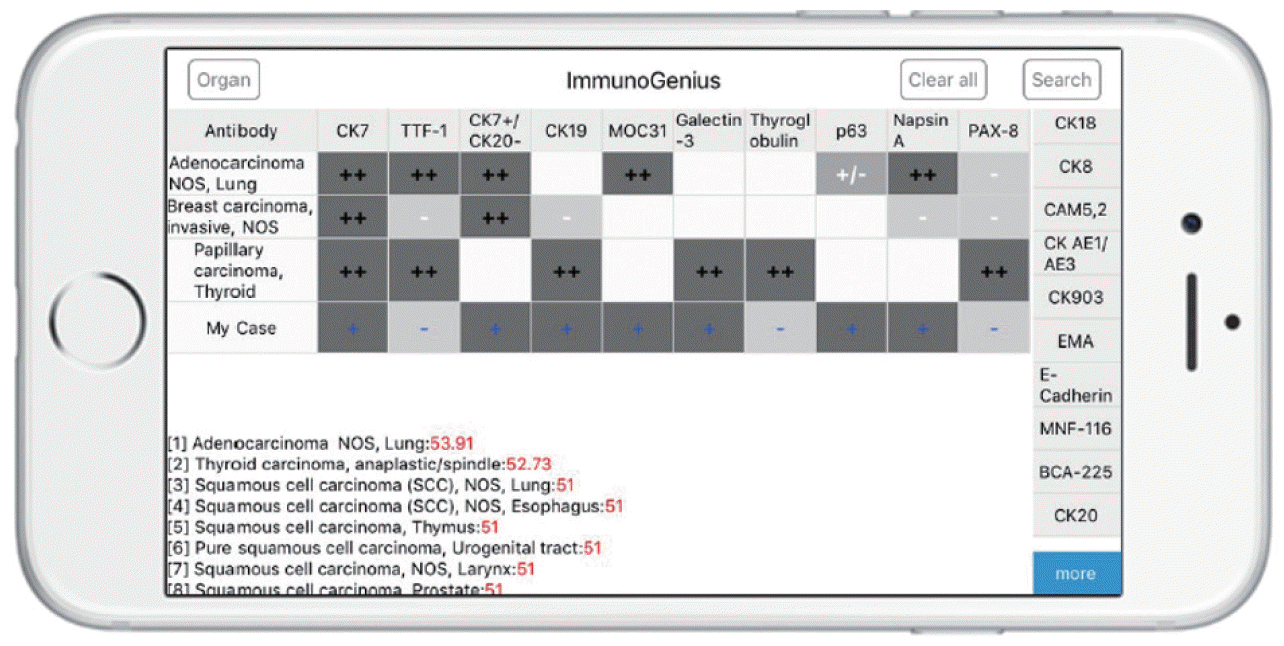
Table 1.
The original diagnosis of the training and validation dataset of lymphomas
|
Original diagnosis |
Training data |
Validation data |
|
B lymphoblastic leukemia/lymphoma |
8 (1.3) |
5 (1.3) |
|
Chronic lymphocytic leukemia/small lymphocytic lymphoma |
20 (3.3) |
11 (2.8) |
|
Extranodal marginal zone lymphoma of MALT (lymphoma) |
78 (13.0) |
74 (18.9) |
|
Nodal marginal zone lymphoma |
4 (0.7) |
5 (1.3) |
|
Plasma cell myeloma |
3 (0.5) |
0 |
|
Follicular lymphoma |
|
|
|
Grade 1, 2, 3A |
59 (9.8) |
16 (4.1) |
|
Grade 3B |
3 (0.5) |
6 (1.5) |
|
Mantle cell lymphoma |
24 (4.0) |
19 (4.8) |
|
DLBCL |
|
|
|
NOS |
216 (34.3) |
145 (37.0) |
|
T-cell/histiocyte-rich |
2 (0.3) |
0 |
|
Primary DLBCL of the CNS |
9 (1.4) |
0 |
|
CD5+ DLBCL |
1 (0.2) |
2 (0.5) |
|
EBV positive DLBCL of elderly |
1 (0.2) |
0 |
|
Anaplastic variant |
0 |
4 (1.0) |
|
Primary cutaneous DLBCL, leg type |
0 |
1 (0.3) |
|
Primary mediastinal (thymic) large B-cell lymphoma |
9 (1.5) |
3 (0.8) |
|
Plasmablastic lymphoma |
3 (0.5) |
0 |
|
Primary effusion lymphoma |
1 (0.2) |
0 |
|
Burkitt lymphoma |
17 (2.8) |
11 (2.8) |
|
B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma |
3 (0.5) |
0 |
|
Primary cutaneous follicle centre lymphoma |
0 |
2 (0.5) |
|
T lymphoblastic leukemia/lymphoma |
17 (2.8) |
7 (1.8) |
|
Extranodal NK/T-cell lymphoma. nasal type |
25 (4.2) |
15 (3.8) |
|
Adult T-cell leukemia/lymphoma |
1 (0.2) |
0 |
|
Enteropathy-associated T-cell lymphoma |
6 (1.0) |
4 (1.0) |
|
Aggressive NK-cell leukemia |
1 (0.2) |
0 |
|
Lymphomatoid papulosis type B |
1 (0.2) |
0 |
|
Mycosis fungoides |
0 |
3 (0.8) |
|
Primary cutaneous (CD30-positive T-cell) ALCL |
1 (0.2) |
0 |
|
Primary cutaneous CD8 positive aggressive epidermotropic cytotoxic T-cell lymphoma |
0 |
2 (0.5) |
|
Subcutaneous panniculitis-like T-cell lymphoma |
0 |
1 (0.3) |
|
Peripheral T-cell lymphoma, NOS |
23 (3.8) |
12 (3.1) |
|
Angioimmunoblastic T-cell lymphoma |
16 (2.7) |
7 (1.8) |
|
ALCL, ALK-positive |
5 (0.8) |
2 (0.5) |
|
ALCL, ALK-negative |
9 (1.5) |
6 (1.5) |
|
Nodular lymphocyte-predominant Hodgkin lymphoma |
0 |
2 (0.5) |
|
Classical Hodgkin lymphoma, NOS |
8 (1.3) |
11 (2.8) |
|
Nodular sclerosis classical Hodgkin lymphoma |
21 (3.5) |
5 (1.3) |
|
Mixed cellularity classical Hodgkin lymphoma |
7 (1.2) |
8 (2.0) |
|
Lymphocyte-depleted classical Hodgkin lymphoma |
0 |
3 (0.8) |
|
Total |
602 (100) |
392 (100) |
Table 2.
Cases with discordant results between the original and predictive diagnosis in training and validation data when searched among lymphomas
|
Original diagnosis |
Error/No. (%)
|
|
Training data |
Validation data |
|
B lymphoblastic leukemia/lymphoma |
0/8 |
0/5 |
|
Chronic lymphocytic leukemia/small lymphocytic lymphoma |
0/20 |
0/11 |
|
Extranodal marginal zone lymphoma of MALT (lymphoma) |
0/78 |
0/74 |
|
Nodal marginal zone lymphoma |
0/4 |
0/5 |
|
Plasma cell myeloma |
0/3 |
0/0 |
|
Follicular lymphoma |
|
|
|
Grade 1, 2, 3A |
0/59 |
0/16 |
|
Grade 3B |
0/3 |
0/6 |
|
Mantle cell lymphoma |
1/24 (4.2) |
0/19 |
|
DLBCL |
|
|
|
NOS |
0/216 |
0/145 |
|
T-cell/histiocyte-rich |
0/2 |
0/0 |
|
Primary DLBCL of the CNS |
0/9 |
0/0 |
|
CD5+ DLBCL |
0/1 |
0/2 |
|
EBV positive DLBCL of elderly |
0/1 |
0/0 |
|
Anaplastic variant |
0/0 |
0/4 |
|
Primary cutaneous DLBCL, leg type |
0/0 |
0/1 |
|
Primary mediastinal (thymic) large B-cell lymphoma |
0/9 |
0/3 |
|
Plasmablastic lymphoma |
3/3 (100) |
0/0 |
|
Primary effusion lymphoma |
0/1 |
0/0 |
|
Burkitt lymphoma |
0/17 |
0/11 |
|
B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma |
0/3 |
0/0 |
|
Primary cutaneous follicle centre lymphoma |
0/0 |
1/2 (50.0) |
|
T lymphoblastic leukemia/lymphoma |
0/17 |
0/7 |
|
Extranodal NK/T-cell lymphoma, nasal type |
0/25 |
0/15 |
|
Adult T-cell leukemia/lymphoma |
1/1 (100) |
0/0 |
|
Enteropathy-associated T-cell lymphoma |
3/6 (50.0) |
2/4 (50.0) |
|
Aggressive NK-cell leukemia |
0/1 |
0/0 |
|
Lymphomatoid papulosis type B |
0/1 |
0/0 |
|
Mycosis fungoides |
0/0 |
0/3 |
|
Primary cutaneous (CD30-positive T-cell) ALCL |
0/1 |
0/0 |
|
Primary cutaneous CD8-positive aggressive epidermotropic cytotoxic T-cell lymphoma |
0/0 |
1/2 (50.0) |
|
Subcutaneous panniculitis-like T-cell lymphoma |
0/0 |
0/1 |
|
Peripheral T-cell lymphoma, NOS |
9/23 (34.7) |
4/12 (33.3) |
|
Angioimmunoblastic T-cell lymphoma |
8/16 (43.8) |
3/7 (42.8) |
|
ALCL, ALK-positive |
0/5 |
0/2 |
|
ALCL, ALK-negative |
3/9 (33.3) |
2/6 (33.3) |
|
Nodular lymphocyte-predominant Hodgkin lymphoma |
0/0 |
1/2 (50.0) |
|
Classical Hodgkin lymphoma, NOS |
1/8 (12.5) |
2/11 (18.2) |
|
Nodular sclerosis classical Hodgkin lymphoma |
3/21 (14.3) |
1/5 (20.0) |
|
Mixed cellularity classical Hodgkin lymphoma |
0/7 |
0/8 |
|
Lymphocyte-depleted classical Hodgkin lymphoma |
0/0 |
1/3 (33.3) |
|
Total |
32/602 (5.3) |
17/392 (4.3) |
Table 3.
A comparison of precision error rates between the training and validation dataset of lymphomas
|
Precision diagnosis |
Training data |
Validation data |
Total |
p-value |
|
Accurate results |
570 (94.7) |
365 (95.7) |
935 (95.0) |
0.543 |
|
Error results |
32 (5.3) |
17 (4.3) |
49 (5.0) |
|
|
Total |
602 (100) |
382 (100) |
984 (100) |
|





 PDF
PDF Citation
Citation Print
Print






 XML Download
XML Download