1. Franklin JM, Schneeweiss S. When and how can real world data analyses substitute for randomized controlled trials? Clin Pharmacol Ther. 2017; 102(6):924–933. PMID:
28836267.

2. Kim HS, Kim JH. Proceed with caution when using real world data and real world evidence. J Korean Med Sci. 2019; 34(4):e28. PMID:
30686950.

3. Kim HS, Lee S, Kim JH. Real-world evidence versus randomized controlled trial: clinical research based on electronic medical records. J Korean Med Sci. 2018; 33(34):e213. PMID:
30127705.

4. Mahmoudi E, Kamdar N, Kim N, Gonzales G, Singh K, Waljee AK. Use of electronic medical records in development and validation of risk prediction models of hospital readmission: systematic review. BMJ. 2020; 369:m958. PMID:
32269037.

5. Kim HS, Kim H, Jeong YJ, Kim TM, Yang SJ, Baik SJ, et al. Development of clinical data mart of HMG-CoA reductase inhibitor for varied clinical research. Endocrinol Metab. 2017; 32(1):90–98.

6. Lee J, Kim TM, Kim H, Lee SH, Cho JH, Lee H, et al. Differences in clinical outcomes between patients with and without hypoglycemia during hospitalization: a retrospective study using real-world evidence. Diabetes Metab J. 2020; 44(4):555–565. PMID:
32431110.

7. Choi J, Bove LA, Tarte V, Choi WJ. Impact of simulated electronic health records on informatics competency of students in informatics course. Healthc Inform Res. 2021; 27(1):67–72. PMID:
33611878.

8. Shin SY. Privacy protection and data utilization. Healthc Inform Res. 2021; 27(1):1–2. PMID:
33611870.

9. Shin SY, Park YR, Shin Y, Choi HJ, Park J, Lyu Y, et al. A de-identification method for bilingual clinical texts of various note types. J Korean Med Sci. 2015; 30(1):7–15. PMID:
25552878.

10. Chevrier R, Foufi V, Gaudet-Blavignac C, Robert A, Lovis C. Use and understanding of anonymization and de-identification in the biomedical literature: scoping review. J Med Internet Res. 2019; 21(5):e13484. PMID:
31152528.

13. Lee D, Park M, Chang S, Ko H. Protecting and utilizing health and medical big data: policy perspectives from Korea. Healthc Inform Res. 2019; 25(4):239–247. PMID:
31777667.

14. Choi HJ, Lee MJ, Choi CM, Lee J, Shin SY, Lyu Y, et al. Establishing the role of honest broker: bridging the gap between protecting personal health data and clinical research efficiency. PeerJ. 2015; 3:e1506. PMID:
26713253.

18. Kim HS, Kim DJ, Yoon KH. Medical big data is not yet available: Why we need realism rather than exaggeration. Endocrinol Metab. 2019; 34(4):349–354.

19. Shin SY. Issues and solutions of healthcare data de-identification: the case of South Korea. J Korean Med Sci. 2018; 33(5):e41. PMID:
29349950.

20. Jones W, Bruce H, Bates MJ, Belkin N, Bergman O, Marshall C. Personal information management in the present and future perfect: reports from a special NSF-sponsored workshop. Proc Am Soc Info Sci Tech. 2005; 42(1):

22. Mandl KD, Perakslis ED. HIPAA and the leak of “deidentified” EHR data. N Engl J Med. 2021; 384(23):2171–2173. PMID:
34110112.

23. Kim H, Baik SY, Yang SJ, Kim TM, Lee SH, Cho JH, et al. Clinical experiences and case review of angiotensin II receptor blocker-related angioedema in Korea. Basic Clin Pharmacol Toxicol. 2019; 124(1):115–122. PMID:
30003686.

24. Mehra MR, Ruschitzka F, Patel AN. Retraction-Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet. 2020; 395(10240):1820. PMID:
32511943.

25. Kim HS, Kim H, Lee H, Park B, Park S, Lee SH, et al. Analysis and comparison of statin prescription patterns and outcomes according to clinical department. J Clin Pharm Ther. 2016; 41(1):70–77. PMID:
26791968.

26. Huh S. Protection of personal information in medical journal publications. Neurointervention. 2019; 14(1):1–8. PMID:
30776876.

27. Galloway A. Estimating actual height in the older individual. J Forensic Sci. 1988; 33(1):126–136. PMID:
3351449.

28. Hartman T, Howell MD, Dean J, Hoory S, Slyper R, Laish I, et al. Customization scenarios for de-identification of clinical notes. BMC Med Inform Decis Mak. 2020; 20(1):14. PMID:
32000770.

29. Purdam K, Elliot M. A case study of the impact of statistical disclosure control on data quality in the individual UK samples of anonymised records. Environ Plan A Econ Space. 2007; 39(5):1101–1118.

30. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972; 18(6):499–502. PMID:
4337382.

31. Nam GE, Park HS. Perspective on diagnostic criteria for obesity and abdominal obesity in Korean adults. J Obes Metab Syndr. 2018; 27(3):134–142. PMID:
31089555.

32. El Sanadi CE, Ji X, Kattan MW. 3-point major cardiovascular event outcome for patients with T2D treated with dipeptidyl peptidase-4 inhibitor or glucagon-like peptide-1 receptor agonist in addition to metformin monotherapy. Ann Transl Med. 2020; 8(21):1345. PMID:
33313090.

33. Hermans WR, Foley DP, Rensing BJ, Rutsch W, Heyndrickx GR, Danchin N, et al. Usefulness of quantitative and qualitative angiographic lesion morphology, and clinical characteristics in predicting major adverse cardiac events during and after native coronary balloon angioplasty. Am J Cardiol. 1993; 72(1):14–20. PMID:
8517422.

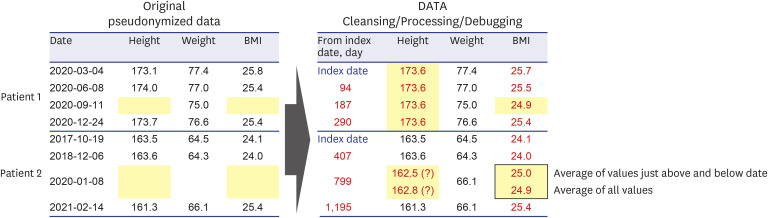
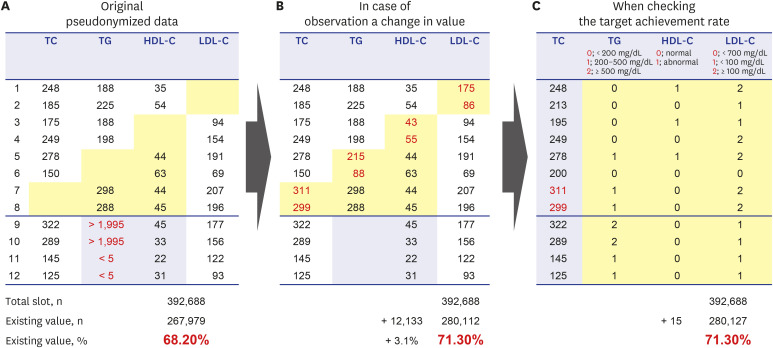
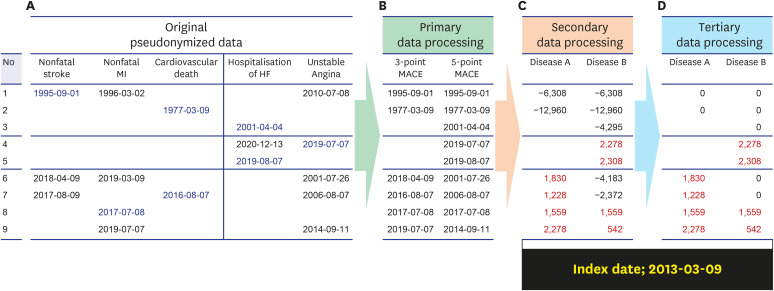




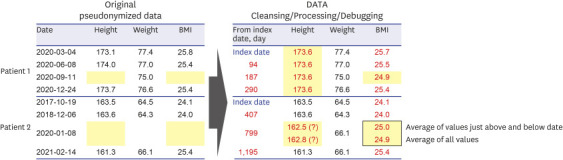
 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download