This article has been corrected. See "Prevalence of a Single-Nucleotide Variant of SARS-CoV-2 in Korea and Its Impact on the Diagnostic Sensitivity of the Xpert Xpress SARS-CoV-2 Assay" in Volume 42 on page 389.
Abstract
The sensitivity of molecular diagnostics could be affected by nucleotide variants in pathogen genes, and the sites affected by such variants should be monitored. We report a single-nucleotide variant (SNV) in the nucleocapsid (N) gene of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), i.e., G29179T, which impairs the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA, USA). We observed significant differences between the threshold cycle (Ct) values for envelope (E) and N genes and confirmed the SNV as the cause of the differences using Sanger sequencing. This SNV, G29179T, is the most prevalent in Korea and is associated with the B.1.497 virus lineage, which is dominant in Korea. Clinical laboratories should be aware of the various SNVs in the SARS-CoV-2 genome and consider their potential effects on the diagnosis of coronavirus disease 2019.
Several PCR primers and probes have been developed for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and various genetic variants have been reported [1-3]. The nucleocapsid (N) gene is a common hotspot for single-nucleotide variants (SNVs), and this has raised concerns about its potential effects on diagnostic sensitivity [1-6]. We report a SNV in N gene that affects the sensitivity of the Xpert Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA, USA). We further evaluated the emergence of this SNV in Korea and found that it is associated with a specific lineage.
The diagnostic performance of the Xpert Xpress assay was retrospectively validated using SARS-CoV-2-positive clinical nasopharyngeal swabs collected from 10 subjects at Seoul Medical Center, Seoul, Korea (Table 1). All specimens were collected in the viral transport medium and were confirmed to be positive by real-time reverse transcription (rRT)-PCR performed using the STANDARD M nCoV Real-Time Detection kit (SD Biosensor, Suwon, Korea) according to the manufacturer’s protocol, and stored at 4°C, from 10 days up to 4 months. As viral RNA is not considered human biological material according to the Enforcement Decree of the Bioethics and Safety Act in Korea, this study was outside the scope of review by an institutional review board and was, therefore, exempted from the requirement of obtaining informed consent from the subjects.
Five specimens were tested using the Xpert Xpress SARS-CoV-2 assay according to the manufacturer’s protocol. Significant differences were observed between the threshold cycle (Ct) values for E and N (Table 1, specimens #6–#10). Particularly, in three specimens, the Ct values for E and N2 differed by 10 cycles, and in the other two specimens, N2 was not detected. The five specimens were retested using both methods, and similar results were obtained.
As nucleotide variants in N may account for the observed Ct value discrepancies, we analyzed the five specimens by Sanger sequencing. No information is available on the exact region in N that is targeted by the Xpert Xpress primers and probes. However, previous reports suggest that the Xpert Xpress assay employs the N2 primer set from the SARS-CoV-2 real-time PCR assay of the United States Centers for Disease Control and Prevention (CDC) [8, 9]. A new primer set (forward, AACACAAGCTTTCGGCAGAC; reverse, TGCAGCAGGAAGAAGAGTCA) was designed to amplify the entire N2 region evaluated in the CDC protocol, with an estimated amplicon size of 385 bp. Sanger sequencing revealed a G>T substitution at position 29,179 (G29179T GenBank: MN908947.3) in all five specimens (Fig. 1). G29179T is located in the 3´-end of the N2 forward primer of the CDC SARS-CoV-2 PCR protocol [10]. No additional SNV was identified in the five specimens.
To confirm that the SNV identified was responsible for the Ct discrepancies, we analyzed five randomly chosen SARS-CoV-2-positive nasopharyngeal swabs (confirmed between March and April 2020 and stored at -70°C) using the Xpert Xpress assay and Sanger sequencing (Table 1, specimens #1–#5). The Ct value discrepancies between E and N were less than four cycles, and Sanger sequencing revealed a G at position 29,179 in each specimen. No other SNV was identified in these additional specimens. These results suggested that G29179T was responsible for the Ct value discrepancies between E and N.
As the exact primer and probe sequences that the Xpert Xpress assay employs are not known—these could be the same, similar, or different from those of the CDC N2 primers—this SNV may differently affect the diagnostic performance of these two methods. However, Ziegler, et al. recently reported a similar phenomenon with the Xpert Xpress assay, which was caused by a different SNV, C29200T, in the N2 region of the CDC primers [7].
To estimate the prevalence of G29179T in Korea, we analyzed 2,489 SARS-CoV-2 sequences from isolates collected in Korea up to February 28, 2021 and stored in the Global Initiative on Sharing All Influenza Data (GISAID) database [1]. In total, 1,740 sequences (69.9%) harbored G29179T. Most G29179T strains belonged to the PANGO lineage B.1.497 (Table 2, Supplemental Data Fig. S1) [11]. Additionally, G29179T has been reported in Peru, Israel, Hong Kong, the United States, Congo, and several other countries, although the prevalence in these countries was significantly lower than that in Korea (Supplemental Data Fig. S2).
The B.1.497 lineage is reported nearly exclusively in Korea and has been designated as a South-Korean lineage [12]. The SNVs that define B.1.497 include C241T, C1059T, C11916T, C14408T, C16560T, A20675T, A23403G, G25563T, and G29179T. This lineage was first reported in Korea on May 5, 2020, and its prevalence has increased rapidly (Table 2) [1]. Although nearly 70% of GISAID sequences from Korea harbor G29179T, genomic surveillance data to accurately estimate the prevalence of B.1.497 in the country are lacking. However, currently, B.1.497 appears to be a predominant lineage in Korea. This finding highlights the need for more extensive genomic surveillance of SARS-CoV-2 in Korea.
The E gene was detected in all specimens. However, in specimens with a low viral load, such as specimens #9 and #10 (Table 1), amplification of E could also be interpreted as a negative result. To date, no issue regarding the diagnostic performance of the Xpert Xpress assay with respect to E has been reported. However, Artesi, et al. reported a SNV in E that causes false-negative results in the Roche Cobas SARS-CoV-2 Test (Roche, Basel, Switzerland) [2]. Additionally, some B.1.497 strains reported in the GISAID database harbor SNVs, such as C26313A, in E [1]. These findings may suggest that some SNVs affecting the detection of E may arise within B.1.497 lineage genomes.
The limitation of our study is that we did not estimate the exact prevalence of B.1.497 lineages, owing to the retrospective nature of this work.
In conclusion, a SNV in N was found to affect the sensitivity of the Xpert Xpress SARS-CoV-2 assay. The SNV identified, G29179T, is the most prevalent in Korea and is associated with the B.1.497 lineage, which is dominant in Korea. Laboratories should be aware of various SNVs in the SARS-CoV-2 genome and their potential effects on the diagnosis of COVID-19.
ACKNOWLEDGEMENTS
We thank Youngkyo Cho (Department of Laboratory Medicine, Yonsei University College of Medicine Severance Hospital) for her contribution to the experiments.
Notes
AUTHOR CONTRIBUTIONS
Hong KH and Lee H take responsibility for the integrity and accuracy of the data. In JW, Lee KA, Kim SH, An YI, and Lee DG performed the PCR and sequencing experiments and analysis. Lee J and Kim SY performed the phylogenetic analysis. Sung H and Kim JS contributed to the revision of the manuscript and the experimental design. All authors have read and approved the final manuscript.
REFERENCES
1. Global initiative on sharing all influenza data (GISAID). Common primer check for high quality genomes. https://www.gisaid.org/hcov-19-analysis-update/. Updated on March 31, 2021.
2. Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, et al. 2020; A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E Gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 58:e01598–20. DOI: 10.1128/JCM.01598-20. PMID: 32690547. PMCID: PMC7512182.

3. Li Z, Li Y, Chen L, Li S, Yu L, Zhu A, et al. 2020; A confirmed case of SARS-CoV-2 pneumonia with routine RT-PCR negative and virus variation in Guangzhou, China. Clin Infect Dis. ciaa941:DOI: 10.1093/cid/ciaa941. PMCID: PMC7454436.
4. Osório NS, Correia-Neves M. 2021; Implication of SARS-CoV-2 evolution in the sensitivity of RT-qPCR diagnostic assays. Lancet Infect Dis. 21:166–7. DOI: 10.1016/S1473-3099(20)30435-7. PMID: 32473662. PMCID: PMC7255733.

5. Álvarez-Díaz DA, Franco-Muñoz C, Laiton-Donato K, Usme-Ciro JA, Franco-Sierra ND, Flórez-Sánchez AC, et al. 2020; Molecular analysis of several in-house rRT-PCR protocols for SARS-CoV-2 detection in the context of genetic variability of the virus in Colombia. Infect Genet Evol. 84:104390. DOI: 10.1016/j.meegid.2020.104390. PMID: 32505692. PMCID: PMC7272177.

6. Vanaerschot M, Mann SA, Webber JT, Kamm J, Bell SM, Bell J, et al. 2020; Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by Reverse Transcription-PCR. J Clin Microbiol. 59:e02369–20. DOI: 10.1128/JCM.02369-20. PMID: 33067272. PMCID: PMC7771441.

7. Ziegler K, Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. 2020; SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 25:2001650. DOI: 10.2807/1560-7917.ES.2020.25.39.2001650. PMID: 33006300. PMCID: PMC7531073.

8. Wolters F, van de Bovenkamp J, van den Bosch B, van den Brink S, Broeders M, Chung NH, et al. 2020; Multi-center evaluation of cepheid xpert xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 128:104426. DOI: 10.1016/j.jcv.2020.104426.

9. Cameron A, Pecora ND, Pettengill MA. 2021; Extraction-free methods for the detection of SARS-CoV-2 by reverse transcription-PCR: a comparison with the Cepheid Xpert Xpress SARS-CoV-2 assay across two medical centers. J Clin Microbiol. 59:e02643–20. DOI: 10.1128/JCM.02643-20. PMID: 33184055. PMCID: PMC8111151.

10. Centers for Disease Convention and Prevention. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Updated on June 6, 2020.
11. Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. 2020; A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 5:1403–7. DOI: 10.1038/s41564-020-0770-5. PMID: 32669681. PMCID: PMC7610519.

12. PANGO Lineages. https://cov-lineages.org/. Updated on March 31, 2021.
Fig. 1
Sanger sequencing results around position 29,179 in the N gene of (A) clinical specimen #5 and (B) clinical specimen #8.
Abbreviation: N, nucleocapsid.
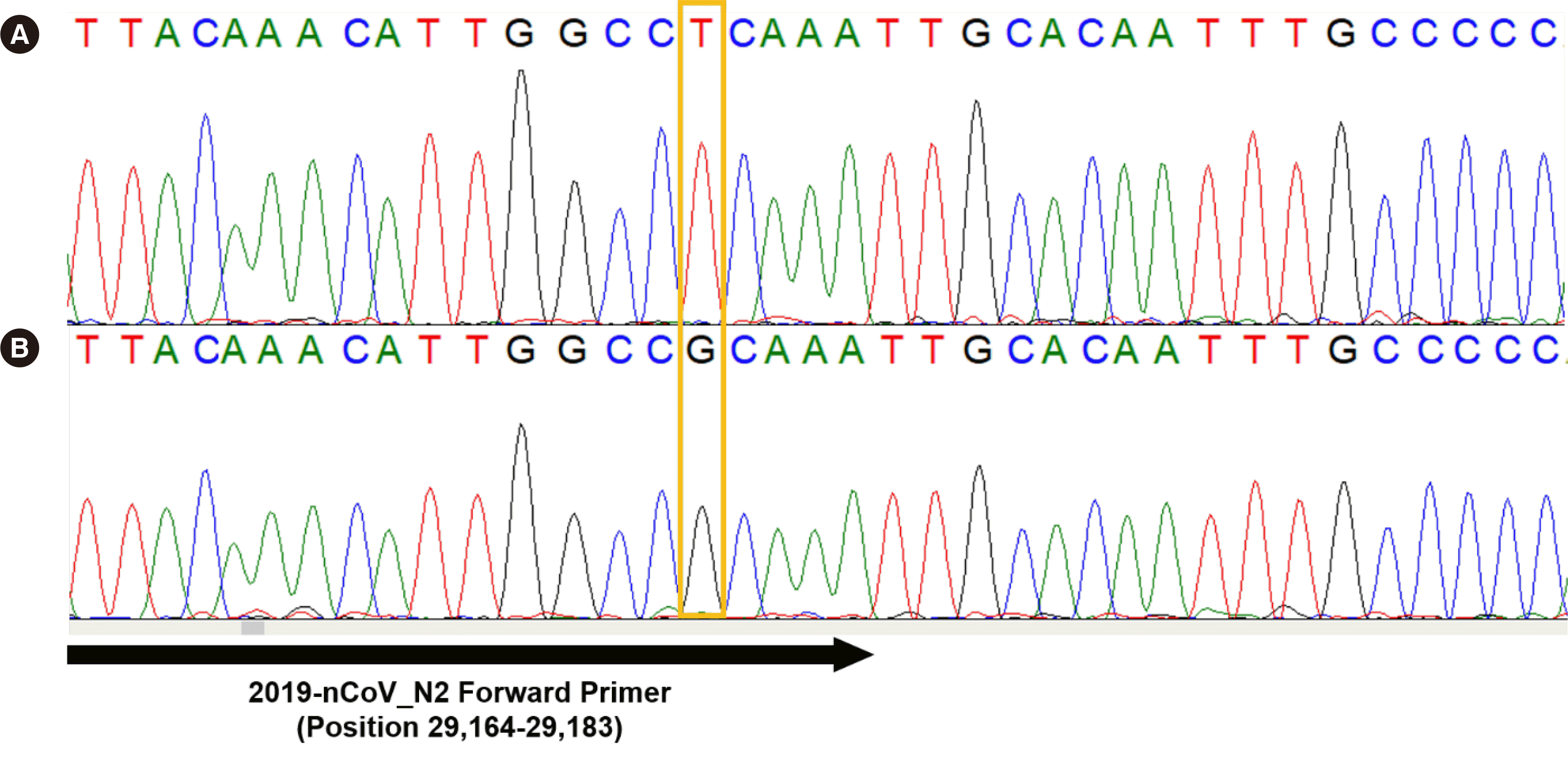
Table 1
Results of rRT-PCR* and Xpert Xpress SARS-CoV-2 assays
Table 2
Emergence of G29179T in SARS-CoV-2 isolates collected in Korea
| Collection | G29179 | T29179 | |
|---|---|---|---|
|
|
|||
| B.1.497* | Others | ||
| Jan 2020 | 8 | ||
| Feb 2020 | 132 | ||
| Mar 2020 | 105 | ||
| Apr 2020 | 23 | ||
| May 2020 | 30 | 172 | |
| Jun 2020 | 40 | 224 | |
| Jul 2020 | 128 | 124 | 4 |
| Aug 2020 | 51 | 242 | 2 |
| Sep 2020 | 28 | 94 | 2 |
| Oct 2020 | 40 | 126 | |
| Nov 2020 | 14 | 239 | 3 |
| Dec 2020 | 26 | 266 | 3 |
| Jan 2021 | 71 | 136 | 1 |
| Feb 2021 | 53 | 102 | |
| Sum | 749 | 1,725 | 15 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download