Abstract
In 2005, the International Council for Harmonization (ICH) established cardiotoxicity assessment guidelines to identify the risk of Torsade de Pointes (TdP). It is focused on the blockade of the human ether-à-go-go-related gene (hERG) channel known to cause QT/QTc prolongation and the QT/QTc prolongation shown on the electrocardiogram. However, these biomarkers are not the direct risks of TdP with low specificity as the action potential is influenced by multiple channels along with the hERG channel. Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative emerged to address limitations of the current model. The objective of CiPA is to develop a standardized in silico model of a human ventricular cell to quantitively evaluate the cardiac response for the cardiac toxicity risk and to come up with a metric for the TdP risk assessment. In silico working group under CiPA developed a standardized and reliable in silico model and a metric that can quantitatively evaluate cellular cardiac electrophysiologic activity. The implementation mainly consists of hERG fitting, Hill fitting, and action potential simulation. In this review, we explained how the in silico model of CiPA works, and briefly summarized current overall CiPA studies. We hope this review helps clinical pharmacologists to understand the underlying estimation process of CiPA in silico modeling.
Since 1986, non-cardiac drugs, mainly terfenadine and astemizole, have been reported to cause QT prolongation and, in some cases, Torsade de Pointes (TdP).[1] Terfenadine and astemizole were withdrawn from the market in 1998 and 1999, respectively, because of the risk of cardiac arrhythmia.[2] Thereafter, the drug-induced QT prolongation has become important in the drug development industry. This issue has been identified as a considerable public health problem and has received attention from the drug regulatory authorities. Regulatory authorities have focused on identifying the risk of TdP during nonclinical and clinical development of a drug.
International Council for Harmonization (ICH) has developed guidelines for safety pharmacology study of a new drug to assess cardiovascular safety to protect patients from the risk of adverse cardiovascular events. In 2005, ICH established guidelines; “The nonclinical evaluation of the potential for delayed ventricular repolarization (QT interval prolongation) by human pharmaceuticals (S7B)” for non-clinical evaluation and “The clinical evaluation of QT/QTc interval prolongation and proarrhythmic potential for non-antiarrhythmic drugs (E14)” for clinical evaluation.[34] These guidelines were recommended for adoption to the regulatory bodies of the European Union, Japan, and the USA. As described in ICH S7B and E14 guidelines, current paradigm to assess the risk of drug-induced TdP is performed on a prolongation of the QTc interval shown on the electrocardiogram (ECG) and blockade of human ether-à-go-go-related gene (hERG) channel, which is considered as one of the important ion channels associated with rapidly activating delayed rectifier potassium current IKr.[5] The implementation of the ICH guidelines has been successful in preventing the introduction of potentially torsadogenic drugs to the market, and thereafter, no drug has been withdrawn from the market due to TdP risk.[6] However, by focusing on hERG blocks and QT prolongation as essential determinants of arrhythmic risk, drug development can be restricted unexpectedly by increasing risk for drugs that may not actually be toxic. More mechanistic-based approach for assessing multichannel interactions was needed to determine the actual TdP proarrhythmic risk.[7]
The Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative, a partnership between Food and Drug Administration (FDA) and several agencies and consortia, including Health Canada, the European Medicines Agency, and Japan's National Institute of Health Sciences, is an effort to overcome limitations of the current assessment model.[8] The objective of CiPA initiative is to facilitate the adoption of a new paradigm for assessment of the clinical potential of TdP that is not measured exclusively by the potency of hERG block and not at all by QT prolongation. Because of the complexity of the task proposed, four work streams that focus on each component of the project, ion channel, in silico, myocyte and early phase human ECG, have been established.[9] For the ion channel work stream, seven important ionic currents playing important roles in generation of action potential were selected - the rapid inward sodium current (INa), the late sodium current (INaL), L-type calcium channel (ICaL), multiple outward potassium currents comprising the transient outward current (Ito), the slow (IKs) and rapid (IKr) components of the delayed rectifier potassium channels, and the inward rectifier channel (IK1)[9] – and voltage clamp protocols for the core set of cardiac ion channel types have been developing. For the in silico workstream, a consensus in silico model has been developed to reconstruct electrophysiologic activity within a heart cell. For the myocyte work stream, capabilities of human stem-cell derived cardiomyocyte assays have been investigated to confirm findings from in vitro and in silico assays. For the early phase human ECG work stream, phase I ECG working group investigated new ECG biomarkers to determine if there are unexpected ion channel effects in humans compared to the preclinical ion channel data.[710]
The CiPA paradigm will be driven by a suite of mechanistically based in vitro assays coupled to in silico reconstructions of cellular cardiac electrophysiologic activity, with verification of completeness through comparison of predicted and observed responses in human-derived cardiomyocytes and early phase human ECG data.[10] Hence, the component of CiPA is to develop a standardized and reliable in silico model and a metric that can quantitatively evaluate cellular cardiac electrophysiologic activity and ultimately assess the risk of cardiotoxicity.
There have been several attempts to assess TdP risk using in silico models, but they were limited to simulating drug effects using the half-maximal blocking concentration (IC50) for different drugs, which assumes simple pore block of the ion channels and neglects any intricacies of drugs ion channel interactions that may be important factors in predicting relative TdP risk.[1112] In CiPA, in silico ventricular action potential (AP) model and a mechanism-based metric for TdP risk stratification were investigated for a more physiologic and pharmacodynamic assessment model. Twenty-eight drugs with well-known characteristics were selected and divided into 12 training drugs and 16 validation drugs for development and validation of an in silico model (Table 1).
The CiPA AP model was developed through a series of modifications to the O'Hara-Rudy (ORd) human ventricular AP model (Fig. 1).[13] A novel hERG/IKr dynamic model was developed by combining a Markov model of the hERG channel that includes temperature-sensitive gating pharmacological components representing open (IO, O) or closed (IC1, IC2, C1, C2) state of hERG channel and a pharmacodynamic model applying drug binding (IO* and O*) and trapping (C*) components (Fig. 2). The hERG/IKr model was then incorporated into the ORd AP model to produce the IKr-dynamic ORd model. Optimization of the model was conducted by scaling ionic current conductances to better reflect changes in AP duration observed in human ventricular myocytes when ionic currents were blocked. The optimized IKr-dynamic ORd model was adopted as a CiPAORdv1.0 model for validation.[14] From this model, in silico biomarker for TdP risk, qNet metric, was derived.[15] As CiPA in silico model aims to assess the integrated effects of multiple ion channel block on TdP risk, uncertainty in drug effects on ion channels to account for variations in experiments is characterized by incorporating uncertainty quantification into modeling predictions.[1516]
Simulating drugs' influence on action potential in human ventricular cells takes three steps: 1) Finding drug binding kinetics parameters of the hERG channel gating model. The non-parametric bootstrap method is used to obtain the joint sampling distribution of drug-hERG parameters. 2) Finding the joint probability distribution comprised of sample's parameter estimation and confidence interval using Markov-chain Monte Carlo (MCMC) simulation in Hill Equation for the remaining six ion currents. 3) Simulating AP in CiPAORdv1.0 model with the joint distributions attained from the previous steps. This software is written in R programming language except for dynamic hERG model and CiPAORdv1.0 model equation parts, which are written in C programming language to improve computing performance. Note that because step 3, AP simulation, uses the results of the prior steps, it must be done after both step 1 and 2 is conducted. Step 1 and 2 are interchangeable.
The dynamic hERG model has 51 parameters. Only five parameters among them are estimated, and the others are fixed. The estimated parameters are Kmax (maximum drug effect at saturating concentrations), Ku (rate of drug unbinding), n (Hill coefficient of drug binding), halfmax (EC50n, nth power of the half-maximal drug concentration), and Vhalf-trap (membrane voltage at which half of the drug-bound channels are open) (Table 2).[17] The hERG model is defined by ordinary differential equations and solved with lsoda solver that selects automatically between stiff and non-stiff methods to solve problems. CMA-ES (Covariance Matrix Adaptation-Evolutionary Strategy) algorithm, which is a stochastic and derivative-free global optimization algorithm for non-linear or non-convex continuous optimization problems, was selected for fitting the hERG model. Model optimization was performed with a population size of 80 and 10−3 of stopping tolerance to minimize objective function value. All parameters to be estimated are encoded logarithmically from their selected ranges [a, b] to the range [0, 10], with the equation below:
The hERG channel in vitro data used to the model fitting should have columns of time, time in milliseconds (ms) during the sweep; frac, fractional current; conc, drug concentration in nM; exp, experiment (cell) number; sweep, the sweep number. A sweep is equivalent of a single pulse of heart and it also equates to one episode in modeling. The data is resampled by non-parametric bootstrapping in order to characterize uncertainty to the parameters of hERG model. In the software developed by Kelly Chang et al., the number of bootstrap samples can be manually set. Prior to performing model fitting for the bootstrap datasets, optimal parameters must be predicted using original data by running the hERG fitting code without any options. The optimal parameter is used as initial values of model parameters for the bootstrap samples.
Objective function used to optimize the model comprises of components comparing differences between observed value and predicted value of the fitted dynamic hERG model. The objective function formula used for model fitting can be described as below.
N is the total number of selected data points across all of doses and episodes. yobs are mean values of bootstrap samples at the specific time points at the same drug concentration and episode, and yPred are predicted values at the same time points of the model fitted by yobs. Small n is the number of examined drug concentrations, and the second and third components in the formula describe the relative reduction of ion current throughout the sweeping procedure to add trapping errors in the function.[17] yObs,rel(1,10) in the second term are relative reductions of the first peak currents between the observed mean values of 1st episode and 10th episode at the specific drug concentrations, and yPred,rel(1,10) are relative reductions of the first peak currents between corresponding simulated values of 1st episode and 10th episode. yObs,rel(1,2) in the third term are relative reductions of the first peak currents between the observed mean values of the 1st episode and 2nd episode at the specific drug concentrations, and yPred,rel(1,2) are relative reductions of the first peak currents between corresponding simulated values of 1st episode and 2nd episode. The two components weigh 0.2*N/conc. Negative error term imposes a penalty to the negative ion current for the negative constraint violation at the same drug concentration.
As the model fitting of several parameters and large samples is computationally intensive, using the computing resource in parallel is recommended. It is the reason for using bootstrap fitting that the method is easier to use parallelization. Optimal parameters and bootstrap parameters for joint sampling distributions are generated as the result data of hERG fitting. A post-process that combines the parameters for an AP simulation step is included in the hERG fitting step.
For non-hERG ionic currents, Hill equation representing the drug-response curve is used for the model. Non-hERG channel in vitro data includes variables of drug names, conc (drug concentration in nM), channel (name of ionic current tested) and block (amount of block (%)). Two parameters IC50 and h (Hill coefficient) are estimated by Hill fitting.
The uncertainty of these parameters is characterized using a Bayesian inference approach. MCMC simulation with the delayed rejection and adaptive Metropolis algorithm is performed to estimate uncertainty in Hill equation parameters using the FME R package.[15] Simulations are performed using model residual function and initial values. The residual function of the model is below:
The initial values are predicted by the optimal nonlinear least squares fit of the Hill equation with the Levenberg-Marquardt algorithm. The initial proposal covariance was set to the scaled parameter covariance matrix of the fitted model. The prior mean of the error variance σ2 (a nuisance parameter) was set to the variance of the fitted residuals, and the prior accuracy parameter was set to give equal weight to the prior and current error variance. By default, the first 10,000 of simulations are discarded as burn-in, and simulation results are saved every 10th iteration over the next 20,000 iterations. The number of burn-in iterations saved simulation results and the intervals can be changed manually. Convergence of parameters is evaluated with the Geweke diagnostic test using version 0.18–1 of coda R package. If the Geweke test comparing the first 10% and the last 50% of the saved iterations indicated a lack of convergence at the 95% confidence interval, then the burn-in for the adaptation period is increased by additional burn-in iterations and the entire MCMC simulation is rerun. This process is repeated until the absolute value of z is greater than 1.96. Optimal parameters and simulated parameters with joint distributions are generated as the result data of Hill fitting.[15]
The CiPAORdv1.0 model is used to simulate action potential, and the base state in the AP simulation code is a slow pacing rate of 2000 ms cycle lengths to mimic bradycardia known as a risk factor for the TdP. If the AP simulation code is run with the default values, it produces action potential at the control states without a drug. Otherwise, defining certain drug name and concentrations without specific sample number can induce simulation with optimal parameters of the drug and concentration. To generate action potential with uncertainty, the number of samples must be defined. Then the sample parameters obtained in the two previous model fitting parts are combined by samples and integrated to simulate action potential. From the simulation results, qNet metric, the net charge carried by ionic currents, are evaluated at each concentration.[18] As this action potential simulation process is computationally intensive similar to the bootstrap fitting process in hERG fitting, it is also recommended to run using the computing resource in parallel.
The qNet metric is computed by integration of the sum of six major currents (IKr, ICaL, INaL, Ito, IKs, and IK1, except INa.) from the start of the stimulus to the end of the beat within the CiPAORdv1.0 model using differential equation solver. [151819] The value of seven important ionic currents were defined as 1 in the default parameter data, so that the membrane voltage and qNet metric can be calculated even if some currents have no in vitro data. After cross-validations to assess TdP risk stratification performance using 12 CiPA training drugs, CiPA in silico working group redefined the optimal metric to assess the TdP risk, as torsade metric score which is an averaged qNet value across 1–4 × free Cmax for each drug with drug effect on the four essential currents – IKr (rapidly activating delayed rectifier potassium current), INaL (late sodium current), ICaL (L-type calcium current), and INa (peak sodium current).[1520]. The measures for model prediction performance were defined as thresholds calculated using data sets of 12 training drugs with ordinal logistic regression. As one of the objectives of CiPA is to use the high throughput patch clamp systems (HTS), “hybrid” data sets collected in both manual and automated were used to validation.[520] Thresholds for the “hybrid” data set were defined by 0.0671 µC/µF for separating low from intermediate/low risk and 0.0581 µC/µF for separating high from intermediate/high risk (Fig. 3).[20]
In vitro studies with human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) and clinical phase I ECG studies were performed to compare whether in silico model integrated with in vitro data well-reflects the response of actual human ventricular cardiomyocytes. In the hiPSC-CM study, sites variability of the assessment of electrophysiology effects of the drugs using either microelectrode array or voltage-sensing optical techniques was characterized, and important hiPSC-CM assay endpoints that had been used to predict drugs for high, intermediate, and low TdP risk categories using linear regression models were identified. The in vitro study was conducted with 28 drugs classified by proarrhythmic risk under the CiPA initiative, using two commercial human cardiomyocyte lines and 5 devices, across the 10 sites. The study showed fairly consistent results across the sites despite the variations in the experimental protocols, and two regression models with the three most useful predictors were constructed predicting the clinical TdP risk well with area under the receiver operating characteristic (ROC) curve values greater than 0.8.[21] The three useful predictors identified in the study were that: 1) ability of a drug to induce “mild” or “severe” arrhythmia-like events at any concentration, 2) the extent of drug-induced repolarization prolongation at any concentration, and 3) the extent of drug-induced prolongation at the clinical Cmax.
The latest clinical phase I ECG study was conducted for validation of an ECG biomarker, a heart rate corrected J-Tpeak (J-Tpeakc) interval, identified at the prior study to differentiate predominant hERG blockers from balanced blockers. For balanced blockers, which is predicted to be low risk by qNet, the CiPA initiative proposes to use ECG analysis in early phase I clinical trials to determine if there is evidence of unexpected ion channel effects in humans compared to the preclinical data. The study showed that concentration-response analysis of QTc and J-Tpeakc can differentiate QTc prolonging drugs with predominant hERG block from drugs that have balanced ion channel block.[22] Results of the latest validation studies suggested that the hiPSC-CM assays can be useful when combined with other CiPA nonclinical assessment strategies, and the ECG data should be interpreted with the nonclinical ion channel data and in silico torsade metric score for more confident assessment. To the extent of the successful validations, the steering team of CiPA is preparing to amend the regulatory requirements for proarrhythmia assessment of the International Council for Harmonization (ICH) S7B and E14 guidelines in compliance with the proposition from ICH S7B/E14 working group.[102324]
The CiPAORdv1.0 model developed under CiPA initiative allows quantitative evaluation of a drug's proarrhythmic risk. Beyond traditional IC50 prediction model confined to hERG/IKr ion channel blockade assessment, the new model is more of a physiologic and pharmacodynamic model that takes account of multiple major ion currents. In vitro data of seven ionic currents – IKr, IKs, ICaL, INaL, Ito, IK1, and INa – is used as model inputs. With the data, in silico model yields a qNet metric which is calculated by six important ionic currents – IKr, IKs, ICaL, INaL, Ito, IK1, except INa. The metric optimized for the assessment of TdP risk is defined as the torsade metric score, which is a mean qNet value averaged across 1–4 × Cmax for each drug with in vitro data of drug effects on the four essential currents – IKr, INa, INaL, and ICaL – as model inputs. Comparisons with the hiPSC-CM assays and the clinical ECG study were conducted for validation of the CiPA in silico model. Eventually, CiPA endeavor will modify the ICH guidelines on assessments of drugs' proarrhythmic risk for more precise and quantitative evaluations.
References
1. Woosley RL, Chen Y, Freiman JP, Gillis RA. Mechanism of the cardiotoxic actions of terfenadine. JAMA. 1993; 269:1532–1536. PMID: 8445816.

2. Li M, Ramos LG. Drug-Induced QT Prolongation And Torsades de Pointes. P T. 2017; 42:473–477. PMID: 28674475.
3. Food and Drug Administration. HHS. International Conference on Harmonisation; guidance on S7B Nonclinical Evaluation of the Potential for Delayed Ventricular Repolarization (QT Interval Prolongation) by Human Pharmaceuticals; availability. Notice. Fed Regist. 2005; 70:61133–61134. PMID: 16237859.
4. Shah RR. Drugs, QTc interval prolongation and final ICH E14 guideline : an important milestone with challenges ahead. Drug Saf. 2005; 28:1009–1028. DOI: 10.2165/00002018-200528110-00003. PMID: 16231954.
5. Sager PT, Gintant G, Turner JR, Pettit S, Stockbridge N. Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am Heart J. 2014; 167:292–300. DOI: 10.1016/j.ahj.2013.11.004. PMID: 24576511.

6. Colatsky T, Fermini B, Gintant G, Pierson JB, Sager P, Sekino Y, et al. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative - Update on progress. J Pharmacol Toxicol Methods. 2016; 81:15–20. DOI: 10.1016/j.vascn.2016.06.002. PMID: 27282641.

7. Vicente J, Zusterzeel R, Johannesen L, Mason J, Sager P, Patel V, et al. Mechanistic Model-Informed Proarrhythmic Risk Assessment of Drugs: Review of the "CiPA" Initiative and Design of a Prospective Clinical Validation Study. Clin Pharmacol Ther. 2018; 103:54–66. DOI: 10.1002/cpt.896. PMID: 28986934.

8. Servick K. A painstaking overhaul for cardiac safety testing. Science. 2016; 353:976–977. PMID: 27701095.

9. Fermini B, Hancox JC, Abi-Gerges N, Bridgland-Taylor M, Chaudhary KW, Colatsky T, et al. A New Perspective in the Field of Cardiac Safety Testing through the Comprehensive In Vitro Proarrhythmia Assay Paradigm. J Biomol Screen. 2016; 21:1–11. DOI: 10.1177/1087057115594589. PMID: 26170255.

10. About CiPA. Accessed 30 September 2018. http://cipaproject.org/about-cipa/.
11. Lancaster MC, Sobie EA. Improved Prediction of Drug-Induced Torsades de Pointes Through Simulations of Dynamics and Machine Learning Algorithms. Clin Pharmacol Ther. 2016; 100:371–379. DOI: 10.1002/cpt.367. PMID: 26950176.

12. Mirams GR, Cui Y, Sher A, Fink M, Cooper J, Heath BM, et al. Simulation of multiple ion channel block provides improved early prediction of compounds' clinical torsadogenic risk. Cardiovasc Res. 2011; 91:53–61. DOI: 10.1093/cvr/cvr044. PMID: 21300721.

13. O'Hara T, Virag L, Varró A, Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011; 7:e1002061. DOI: 10.1371/journal.pcbi.1002061. PMID: 21637795.
14. github.com/FDA/CiPA. Accessed 30 September 2018. https://github.com/FDA/CiPA.
15. Chang KC, Dutta S, Mirams GR, Beattie KA, Sheng J, Tran PN, et al. Uncertainty Quantification Reveals the Importance of Data Variability and Experimental Design Considerations for in Silico Proarrhythmia Risk Assessment. Front Physiol. 2017; 8:917. DOI: 10.3389/fphys.2017.00917. PMID: 29209226.

16. Pathmanathan P, Shotwell MS, Gavaghan DJ, Cordeiro JM, Gray RA. Uncertainty quantification of fast sodium current steady-state inactivation for multi-scale models of cardiac electrophysiology. Prog Biophys Mol Biol. 2015; 117:4–18. DOI: 10.1016/j.pbiomolbio.2015.01.008. PMID: 25661325.

17. Li Z, Dutta S, Sheng J, Tran PN, Wu W, Chang K, et al. Improving the In Silico Assessment of Proarrhythmia Risk by Combining hERG (Human Ether-a-go-go-Related Gene) Channel-Drug Binding Kinetics and Multichannel Pharmacology. Circ Arrhythm Electrophysiol. 2017; 10:e004628. DOI: 10.1161/circep.116.004628. PMID: 28202629.

18. Dutta S, Chang KC, Beattie KA, Sheng J, Tran PN, Wu WW, et al. Optimization of an In silico Cardiac Cell Model for Proarrhythmia Risk Assessment. Front Physiol. 2017; 8:616. DOI: 10.3389/fphys.2017.00616. PMID: 28878692.

19. In Silico Proarrhythmia Risk Assessment under the CiPA Initiative. Accessed 30 September 2018. http://cipaproject.org/wp-content/uploads/sites/24/2017/11/FINALSPS2017AnnualMeeting_CiPAModeling.pdf.
20. Li Z, Ridder BJ, Han X, Wu WW, Sheng J, Tran PN, et al. Assessment of an In Silico Mechanistic Model for Proarrhythmia Risk Prediction Under the CiPA Initiative. Clin Pharmacol Ther. 2019; 105:466–475. DOI: 10.1002/cpt.1184. PMID: 30151907.
21. Blinova K, Dang Q, Millard D, Smith G, Pierson J, Guo L, et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep. 2018; 24:3582–3592. DOI: 10.1016/j.celrep.2018.08.079. PMID: 30257217.

22. Vicente J, Zusterzeel R, Johannesen L, Ochoa-Jimenez R, Mason JW, Sanabria C, et al. Assessment of Multi-Ion Channel Block in a Phase-1 Randomized Study Design: Results of the CiPA Phase 1 ECG Biomarker Validation Study. Clin Pharmacol Ther. 2018; DOI: 10.1002/cpt.1303.
23. Final Concept Paper - ICH S7B and E14 Q&A. Accessed 10 March 2019. https://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Efficacy/E14/E14S7BIWG_ConceptPaper_Final_2018_1122.pdf.
24. Strauss DG, Gintant G, Li Z, Wu W, Blinova K, Vicente J, et al. Comprehensive In Vitro Proarrhythmia Assay (CiPA) Update from a Cardiac Safety Research Consortium / Health and Environmental Sciences Institute / FDA Meeting. Ther Innov Regul Sci. 2018; 2168479018795117. DOI: 10.1177/2168479018795117.

Figure 1
Schematic diagram of O'Hara-Rudy human ventricular myocyte model. Among various ion currents in the model, in silico studies in CiPA focus on the seven major ion currents: IKr, IKs, ICaL, INaL, INa, Ito and IK1. IKr/hERG, rapid delayed rectifier potassium current that flows through0 the hERG channel; IKs, the slow rectifier potassium current; ICaL, the L-type calcium current; INa, the peak sodium current; INaL, the late sodium current; Ito, transient outward potassium current; IK1, inwardly rectifying potassium current. Adapted from ref. 13.
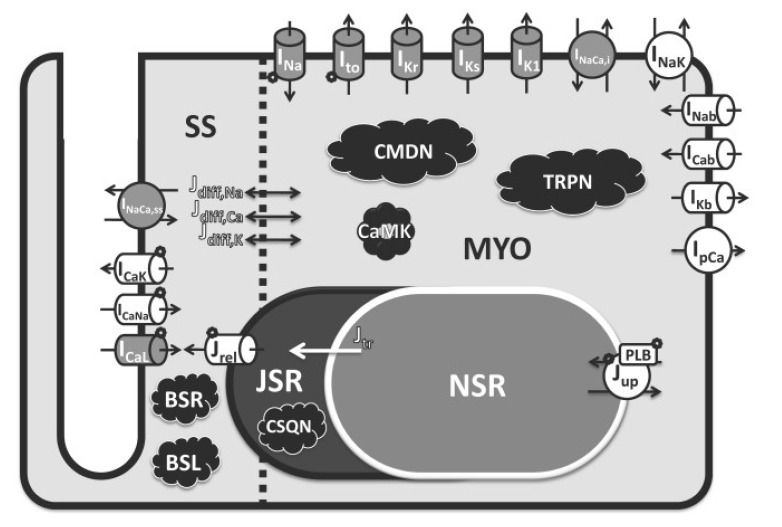
Figure 2
Structure of the hERG Markov model and equation of transition rate affected by temperature. The transition between adjacent states is a first order reaction dependent on membrane voltage, temperature and three free parameters A, B, and q. Each state transition has a different set of free parameters that are fixed and distinguished from each other by numeric suffixes. R, state transition rate; V, membrane potential (electrical field across the channel); A and B, energy barrier height in the absence and presence of electrical field, respectively; T, temperature; q, commonly used temperature extrapolating Q10 value defined as the change in rate for each 10℃ change in temperature. The hERG Markov model in CiPA in silico assay includes a saturating drug binding component (IO* and O*) and a drug trapping component (C*). The fitted drug parameters are Ku (drug unbinding rate), Kmax (maximum drug effect), n (Hill coefficient of drug binding), halfmax (EC50n, nth power of the half-maximal drug concentration), and Vhalf-trap (membrane voltage of half of drug-bound channels opening). Emax is a sigmoid model describing the concentration-response of each drug and D is the drug concentration in nM containing. The trapping rate (Kt) of channel closing with drug bound was manually fixed at 3.5×10−5 ms−1. Modified from ref. 14 and ref. 17.
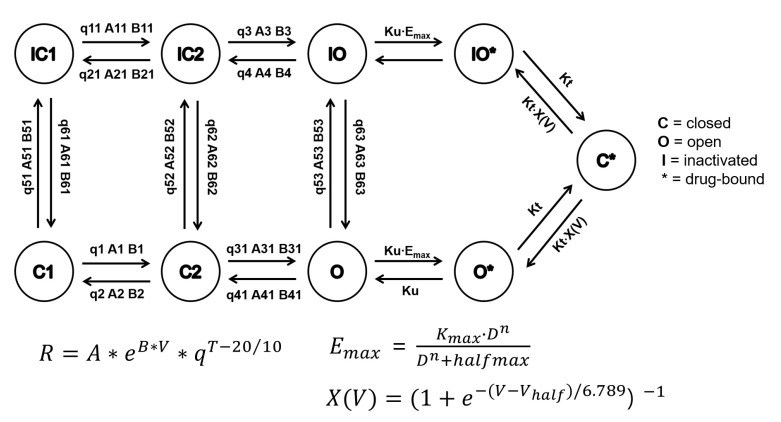
Figure 3
The finalized TdP risk metric of average qNet for 1–4 × Cmax of 28 CiPA drugs. Results shown are for the 12 training drugs (a) and 16 validation drugs (b), respectively. The 95% confidence interval and median point of the torsade metric scores for each drug are shown as horizontal error bars. Two dotted lines represent the thresholds of the risk category (Threshold 2 has a value of 0.0581 and Threshold 1 has a value of 0.0671). The metric was plotted using data obtained from “hybrid” data set which combined manual data of hERG current with high-throughput data of the other three currents (INaL, INa, and ICaL). The data was provided by Dr. Zhihua Li at FDA.
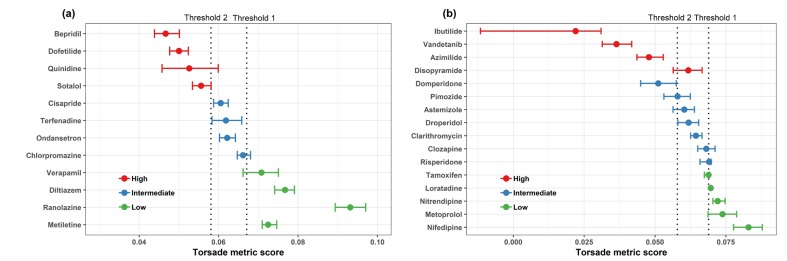
Table 1
CiPA drugs selected for model development by the clinical transitional working group. The drugs are divided into 12 training drugs and 16 validation drugs with well-known Torsade de Pointes risk (high/intermediate/low risk)
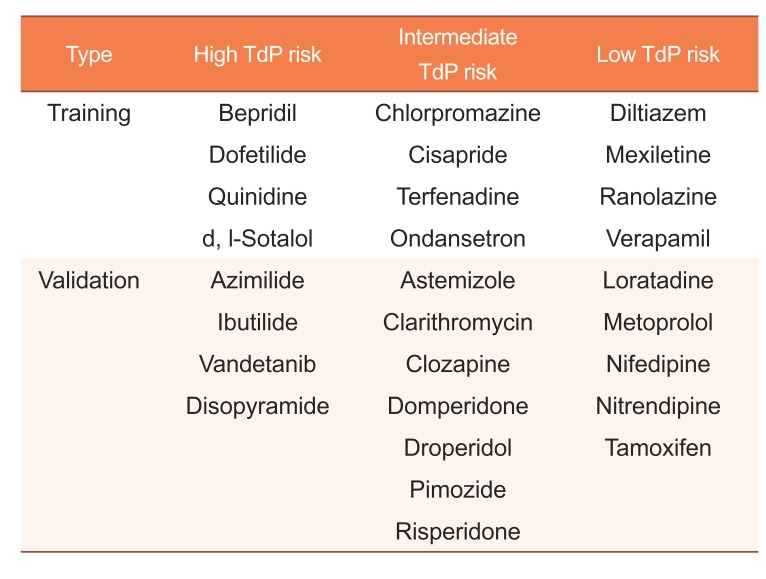
Table 2
Parameters of dynamic hERG model. A total of 51 parameters with 46 fixed parameters (Five parameters in bold style are estimated in hERG fitting step)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


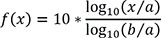
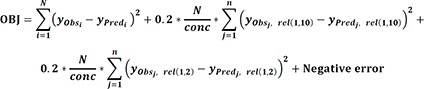
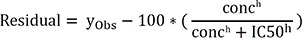
 XML Download
XML Download