1. Ueda SM, Kapp DS, Cheung MK, Shin JY, Osann K, Husain A, et al. Trends in demographic and clinical characteristics in women diagnosed with corpus cancer and their potential impact on the increasing number of deaths. Am J Obstet Gynecol. 2008; 198:218.e1–218.e6.
2. Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012; 13:679–692.
3. Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005; 6:597–610.
4. Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000; 24:132–138.
5. Yao L, Ren S, Zhang M, Du F, Zhu Y, Yu H, et al. Identification of specific DNA methylation sites on the Y-chromosome as biomarker in prostate cancer. Oncotarget. 2015; 6:40611–40621.
6. Huang RL, Su PH, Liao YP, Wu TI, Hsu YT, Lin WY, et al. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017; 23:263–272.
7. Sheng Y, Wang H, Liu D, Zhang C, Deng Y, Yang F, et al. Methylation of tumor suppressor gene CDH13 and SHP1 promoters and their epigenetic regulation by the UHRF1/PRMT5 complex in endometrial carcinoma. Gynecol Oncol. 2016; 140:145–151.
8. Chen YC, Tsao CM, Kuo CC, Yu MH, Lin YW, Yang CY, et al. Quantitative DNA methylation analysis of selected genes in endometrial carcinogenesis. Taiwan J Obstet Gynecol. 2015; 54:572–579.
9. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559.
10. Amrine KC, Blanco-Ulate B, Cantu D. Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PLoS One. 2015; 10:e0118731.
11. Busch R, Qiu W, Lasky-Su J, Morrow J, Criner G, DeMeo D. Differential DNA methylation marks and gene comethylation of COPD in African-Americans with COPD exacerbations. Respir Res. 2016; 17:143.
12. Jia N, Wang J, Li Q, Tao X, Chang K, Hua K, et al. DNA methylation promotes paired box 2 expression via myeloid zinc finger 1 in endometrial cancer. Oncotarget. 2016; 7:84785–84797.
13. Trimarchi MP, Yan P, Groden J, Bundschuh R, Goodfellow PJ. Identification of endometrial cancer methylation features using combined methylation analysis methods. PLoS One. 2017; 12:e0173242.
14. Zhang B, Xing X, Li J, Lowdon RF, Zhou Y, Lin N, et al. Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC Genomics. 2014; 15:868.
15. Warden CD, Lee H, Tompkins JD, Li X, Wang C, Riggs AD, et al. COHCAP: an integrative genomic pipeline for single-nucleotide resolution DNA methylation analysis. Nucleic Acids Res. 2013; 41:e117.
16. Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014; e005165.
17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550.
18. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504.
19. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010; 28:495–501.
20. Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012; 40:W478-83.
21. Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA. 2005; 102:3336–3341.
22. Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997; 13:335–340.
23. Piulats JM, Guerra E, Gil-Martín M, Roman-Canal B, Gatius S, Sanz-Pamplona R, et al. Molecular approaches for classifying endometrial carcinoma. Gynecol Oncol. 2017; 145:200–207.
24. Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008; 27:4657–4665.
25. Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, et al. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010; 31:1027–1036.
26. Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, et al. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007; 67:2617–2625.
27. Wang MX, Wang HY, Zhao X, Srilatha N, Zheng D, Shi H, et al. Molecular detection of B-cell neoplasms by specific DNA methylation biomarkers. Int J Clin Exp Pathol. 2010; 3:265–279.
28. Waha A, Güntner S, Huang TH, Yan PS, Arslan B, Pietsch T, et al. Epigenetic silencing of the protocadherin family member PCDH-gamma-A11 in astrocytomas. Neoplasia. 2005; 7:193–199.
29. Dallosso AR, Hancock AL, Szemes M, Moorwood K, Chilukamarri L, Tsai HH, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 2009; 5:e1000745.
30. Divine LM, Nguyen MR, Meller E, Desai RA, Arif B, Rankin EB, et al. AXL modulates extracellular matrix protein expression and is essential for invasion and metastasis in endometrial cancer. Oncotarget. 2016; 7:77291–77305.
31. Du XL, Jiang T, Zhao WB, Wang F, Wang GL, Cui M, et al. Gene alterations in tumor-associated endothelial cells from endometrial cancer. Int J Mol Med. 2008; 22:619–632.
32. Lévy P, Ripoche H, Laurendeau I, Lazar V, Ortonne N, Parfait B, et al. Microarray-based identification of tenascin C and tenascin XB, genes possibly involved in tumorigenesis associated with neurofibromatosis type 1. Clin Cancer Res. 2007; 13:398–407.
33. Hsu MK, Wu IC, Cheng CC, Su JL, Hsieh CH, Lin YS, et al. Triple-layer dissection of the lung adenocarcinoma transcriptome: regulation at the gene, transcript, and exon levels. Oncotarget. 2015; 6:28755–28773.
34. Margolin G, Petrykowska HM, Jameel N, Bell DW, Young AC, Elnitski L. Robust detection of DNA hypermethylation of ZNF154 as a pan-cancer locus with in silico modeling for blood-based diagnostic development. J Mol Diagn. 2016; 18:283–298.
35. Sánchez-Vega F, Gotea V, Petrykowska HM, Margolin G, Krivak TC, DeLoia JA, et al. Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics. 2013; 8:1355–1372.
36. Saied MH, Marzec J, Khalid S, Smith P, Down TA, Rakyan VK, et al. Genome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeats. PLoS One. 2012; 7:e33213.


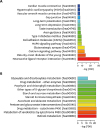





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download