Abstract
Objective
We wanted to investigate the mechanisms that could account for the pathogenesis of rheumatoid arthritis, so we examined the different expressions of the genes in rheumatoid arthritis (RA) synovial fluid macrophages as compared with that of normal peripheral blood (PB) mon-ocyte-derived macrophages using microarray and bioinformatic analysis.
Methods
We examined the expression of genes by using a gene expression oligonucleotide microarray. The differences of the gene expressions between the RA synovial macrophages and the normal PB monocytes-derived macrophages were analyzed using bioinformatic tools, including cytoscape and its plugin.
Results
In this study, we found that 899 genes (464 genes upregulated and 435 genes down-regulated) were differentially expressed between the two groups. Among the 899 genes, 552 genes were included for gene ontology analysis and network analysis. Based on biological process ontology, they were categorised mainly into immune response processes, responses to stimulus and signaling and regulation of biological processes. In addition to the genes related with STAT1 and AP-1 signaling, we found that the genes involved in the antigen processing and the cell cycle are abundantly expressed in RA synovial macrophages, sug-gesting that these genes may play an important role in the pathogenesis of RA.
Go to : 
References
2. Teixeira VH, Olaso R, Martin-Magniette ML, Lasbleiz S, Jacq L, Oliveira CR, et al. Transcriptome analysis de-scribing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One. 2009; 4:e6803.

3. Toonen EJ, Barrera P, Radstake TR, van Riel PL, Scheffer H, Franke B, et al. Gene expression profiling in rheumatoid arthritis: current concepts and future directions. Ann Rheum Dis. 2008; 67:1663–9.

4. van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007; 66:1008–14.

5. van Baarsen LG, Bos CL, van der Pouw Kraan TC, Verweij CL. Transcription profiling of rheumatic diseases. Arthritis Res Ther. 2009; 11:207.

6. Bansard C, Lequerré T, Derambure C, Vittecoq O, Hiron M, Daragon A, et al. Gene profiling predicts rheumatoid arthritis responsiveness to IL-1Ra (anakinra). Rheumatology (Oxford). 2011; 50:283–92.

7. Lindberg J, Wijbrandts CA, van Baarsen LG, Nader G, Klareskog L, Catrina A, et al. The gene expression profile in the synovium as a predictor of the clinical response to infliximab treatment in rheumatoid arthritis. PLoS One. 2010; 5:e11310.

8. Diez D, Wheelock AM, Goto S, Haeggström JZ, Paulsson-Berne G, Hansson GK, et al. The use of network analyses for elucidating mechanisms in cardiovascular disease. Mol Biosyst. 2010; 6:289–304.

9. Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003; 34:166–76.

10. Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007; 2:2366–82.

11. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–504.

12. Antoniv TT, Ivashkiv LB. Dysregulation of interleukin-10-dependent gene expression in rheumatoid arthritis synovial macrophages. Arthritis Rheum. 2006; 54:2711–21.

13. Huang Q, Ma Y, Adebayo A, Pope RM. Increased macrophage activation mediated through toll-like receptors in rheumatoid arthritis. Arthritis Rheum. 2007; 56:2192–201.

14. Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005; 6:R40.
15. Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005; 437:1173–8.

16. Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005; 122:957–68.

17. van der Pouw Kraan TC, van Gaalen FA, Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003; 48:2132–45.

18. Haehnel V, Schwarzfischer L, Fenton MJ, Rehli M. Transcriptional regulation of the human toll-like receptor 2 gene in monocytes and macrophages. J Immunol. 2002; 168:5629–37.
19. Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004; 279:7370–7.

20. Taniguchi N, Kawahara K, Yone K, Hashiguchi T, Yamakuchi M, Goto M, et al. High mobility group box chromosomal protein 1 plays a role in the pathogenesis of rheumatoid arthritis as a novel cytokine. Arthritis Rheum. 2003; 48:971–81.

21. Xu W, Comhair SA, Zheng S, Chu SC, Marks-Konczalik J, Moss J, et al. STAT-1 and c-Fos interaction in nitric oxide synthase-2 gene activation. Am J Physiol Lung Cell Mol Physiol. 2003; 285:L137–48.
22. Rivett AJ, Bose S, Brooks P, Broadfoot KI. Regulation of proteasome complexes by gamma-interferon and phosphorylation. Biochimie. 2001; 83:363–6.
23. Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukoc Biol. 1994; 56:571–5.
24. Nonomura Y, Kohsaka H, Nagasaka K, Miyasaka N. Gene transfer of a cell cycle modulator exerts antiinflammatory effects in the treatment of arthritis. J Immunol. 2003; 171:4913–9.

25. Perlman H, Bradley K, Liu H, Cole S, Shamiyeh E, Smith RC, et al. IL-6 and matrix metalloproteinase-1 are regulated by the cyclin-dependent kinase inhibitor p21 in synovial fibroblasts. J Immunol. 2003; 170:838–45.

26. Scatizzi JC, Hutcheson J, Bickel E, Woods JM, Klosowska K, Moore TL, et al. p21Cip1 is required for the development of monocytes and their response to serum transfer-induced arthritis. Am J Pathol. 2006; 168:1531–41.

27. Kwak HB, Jin HM, Ha H, Kang MJ, Lee SB, Kim HH, et al. Tumor necrosis factor-alpha induces differentiation of human peripheral blood mononuclear cells into osteoclasts through the induction of p21(WAF1/Cip1). Biochem Biophys Res Commun. 2005; 330:1080–6.
28. Sankar U, Patel K, Rosol TJ, Ostrowski MC. RANKL coordinates cell cycle withdrawal and differentiation in osteoclasts through the cyclin-dependent kinase inhibitors p27KIP1 and p21CIP1. J Bone Miner Res. 2004; 19:1339–48.
29. Perlman H, Pagliari LJ, Liu H, Koch AE, Haines GK 3rd, Pope RM. Rheumatoid arthritis synovial macrophages express the Fas-associated death domain-like in-terleukin-1beta-converting enzyme-inhibitory protein and are refractory to Fas-mediated apoptosis. Arthritis Rheum. 2001; 44:21–30.
Go to : 
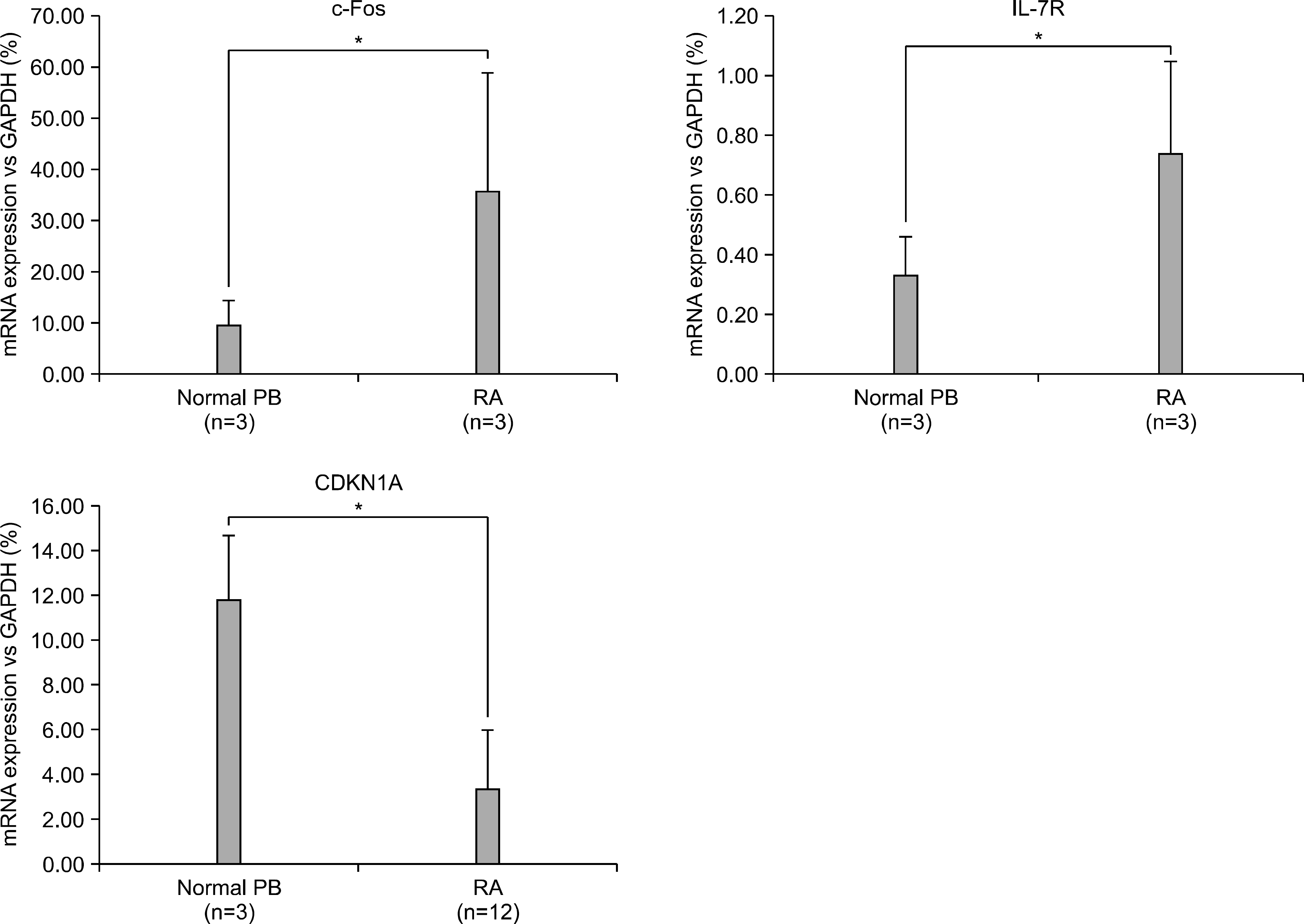 | Figure 2.The array data for the gene expressions was validated by performing quantitative real-time PCR in the healthy volunteer PB monocyte-derive macrophages and the RA synovial macrophages ∗p<0.05 versus the healthy volunteer PB monocyte-derive macrophages. |
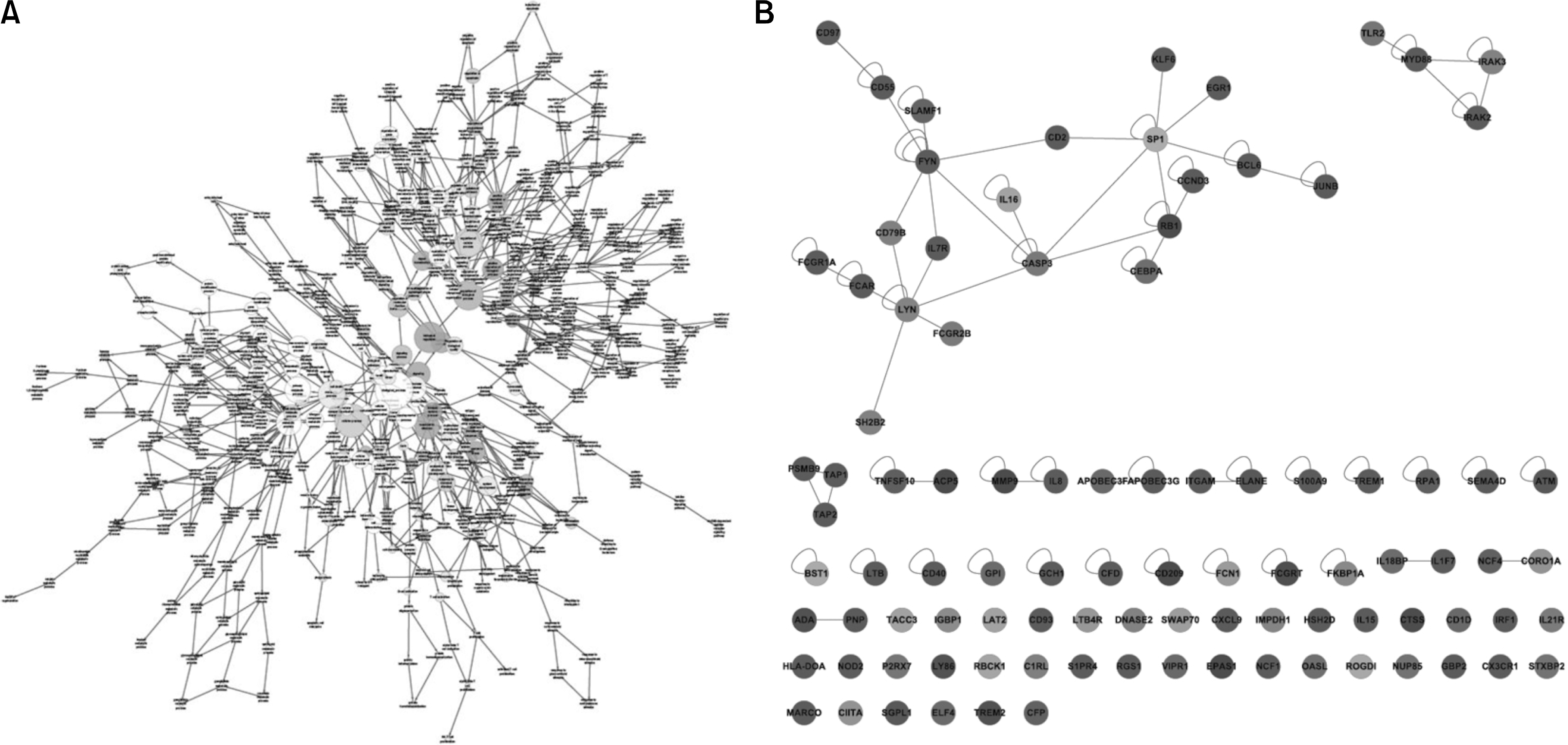 | Figure 3.(A) Map of the biological processes associated with RA synovial macrophages. Darker nodes mean the more significant ontology terms. The size is proportional to the number of genes included in that ontology term. (B) Network of the genes included in the immune system processes (GO. 2376). A blue node means down-regulation of genes and a red node means upregulation of genes in the RA synovial macrophages. |
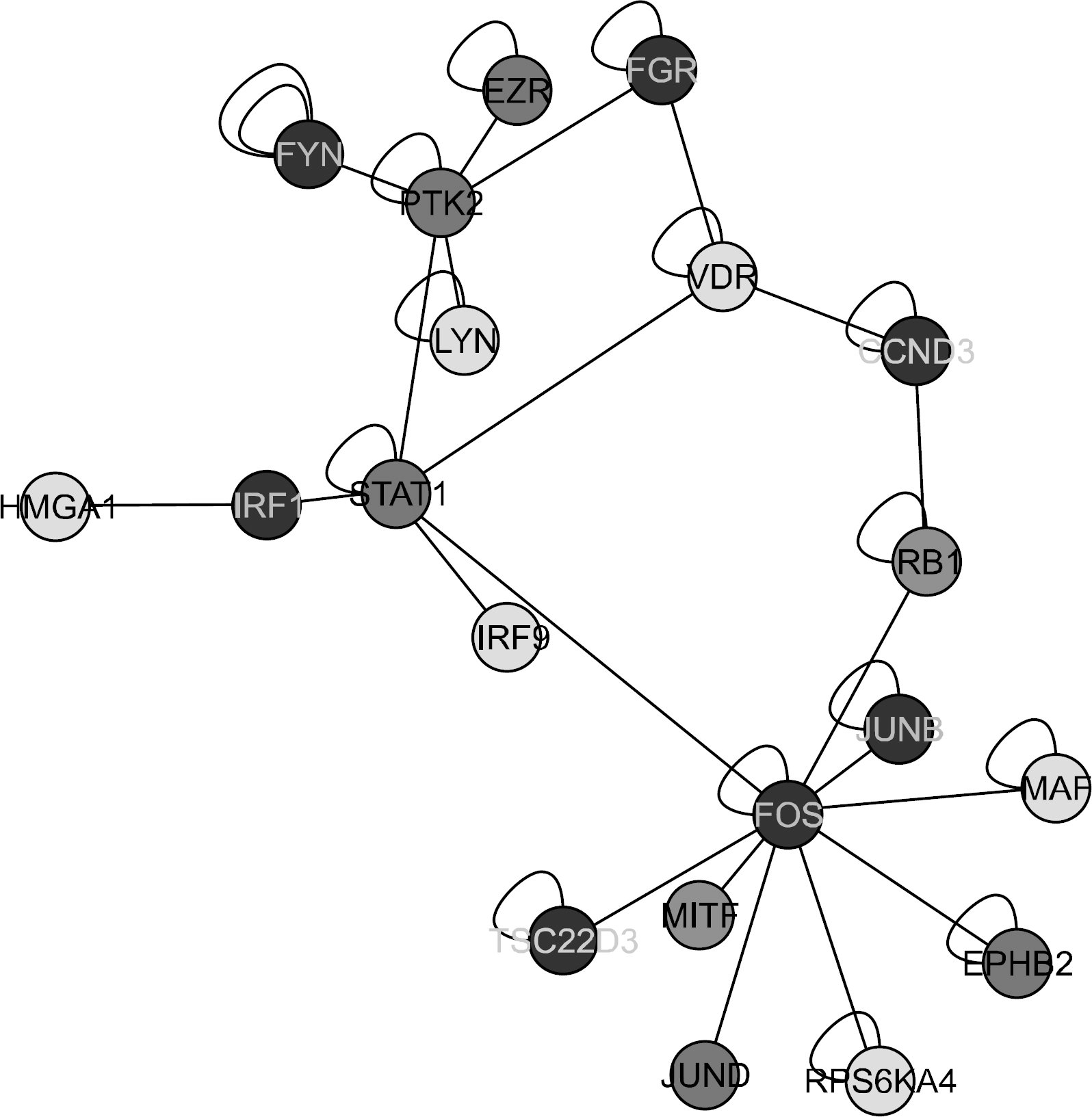 | Figure 4.Different expressions of the STAT1-related genes in the RA synovial macrophages. A blue node means down-regulation of genes and a red node means upregulation of genes in the RA synovial macrophages. |
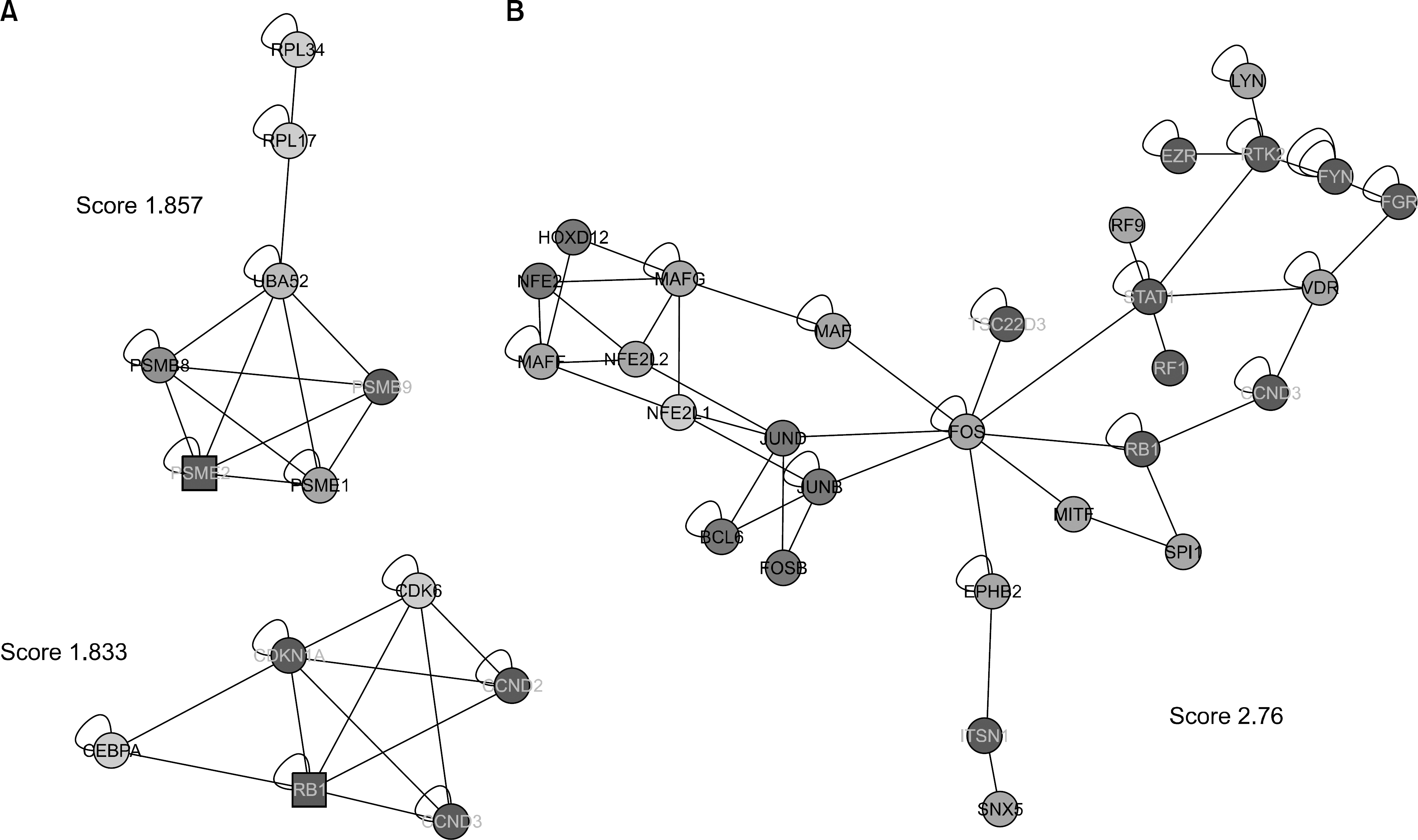 | Figure 5.(A) Detection of densely connected regions in the network of genes differentially expressed in the RA synovial macrophages and the PB monocyte-derived macrophages from healthy volunteer using MCODE. A square node is a seed node. (B) Identification of the functional modules as highly connected regions with similar responses using jActiveModules. A blue node means down-regulation of genes and a red node means upregulation of genes in the RA synovial macrophages. |
Table 1.
The gene ontology analysis (biological process) of the differentially expressed genes in RA synovial macrophages using the BiNGO plugin (The top ten GO terms were statistically significant)
Table 2.
The gene ontology analysis (molecular function) of the differentially expressed genes in RA synovial macrophages using the BiNGO plugin (The top ten GO terms were statistically significant)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download