Abstract
Cadmium intoxication has been associated with the dysregulation of iron homeostasis. In the present study, we investigated the effect of cadmium on the expression of ferroportin 1 (FPN1), an important iron transporter protein that is involved in iron release from macrophages. When we incubated cadmium with J774 mouse macrophage cells, FPN1 mRNA levels were significantly increased in a dose- and time-dependent manner. Furthermore, the cadmium-induced FPN1 mRNA expression was associated with increased levels of FPN1 protein. On the other hand, cadmium-mediated FPN1 mRNA induction in J774 cells was completely blocked when cells were co-treated with a transcription inhibitor, acitomycin D. Also, cadmium directly stimulated the activity of the FPN1-promoter driven luciferase reporter, suggesting that the cadmium up-regulates FPN1 gene expression in a transcription-dependent manner. Finally, cadmium exposure to J774 macrophages increased intracellular reactive oxygen species (ROS) levels by ~ 2-fold, compared to untreated controls. When J774 cells were co-treated with antioxidant N-acetylcystein, the cadmium-induced FPN1 mRNA induction was significantly attenuated. In summary, the results of this study clearly demonstrated that cadmium increased FPN1 expression in macrophages through a mechanism that involves ROS production, and suggests another important interaction between iron and cadmium metabolism.
Cadmium is a well-known toxic metal widely distributed in the environment. Cadmium compounds are present in various products such as plastic stabilizers, color pigment, several alloys, certain batteries, and phosphate fertilizers (Järup, 2003). Cadmium intoxication in the general population can occur through cigarette smoking, habitual intake of contaminated food or water and by inhalation of polluted air (WHO, 2000). Each cigarette contains approximately 1~2 ug of cadmium, with 40-60% of that being absorbed through the lungs directly into the systemic circulation (ATSDR, 2003). The major contaminated sources of dietary cadmium are fish, liver, grains, leafy vegetables, potatoes, and other root vegetables. Once absorbed, cadmium has a long biological half-life of 10 to 30 years and its excretion from the body is extremely low (Satarug et al., 2003). Therefore, it accumulates in various tissues such as kidneys, lung, liver, bone, testis and placenta (Djukic-Cosic et al., 2008). Studies have shown that chronic cadmium exposure even at low levels leads to a variety of illness including kidney dysfunction, osteoporosis, early onset of diabetic renal complications, hypertension, and increased cancer risk (Järup et al., 1998; Staessen et al., 1996). The metal and its compounds are in fact classified as group 1 carcinogens for humans (IARC, 1993).
Studies on cadmium toxicity have revealed diverse cellular targets for the deleterious action of this toxic metal. Among them, growing evidence has accumulated that cadmium perturbs the homeostasis of essential metals, especially iron. For example, the production of erythropoietin, an important regulator of iron metabolism in the body, is markedly decreased in cadmium poisoned kidneys (Horiguchi, 2007; Schafer et al., 1990). Furthermore, other important steps of iron homeostasis, such as iron absorption from the diet, contribute to cadmium toxicity. In the case of iron deficiency, production of divalent metal transporter 1 (DMT1), an iron importer that is located in the apical membrane of enterocytes, is significantly increased (Gunshin et al., 1997). Interestingly, the induction of DMT1 was shown to be associated with increased cadmium burden in mice fed an iron-deficient diet (Ryu et al., 2004). DMT1 can transport several other divalent metal ions in addition to iron, including cadmium (Bressler et al., 2004). Therefore, investigations on the role of cadmium in the regulation of iron transporters would be useful to better understand the interactions between iron and cadmium metabolism.
Macrophages play a key role in maintaining body iron homeostasis. Especially, macrophages of reticuloendothelial system, such as Kupffer cells in the liver and macrophages of spleen and bone marrow, provide most of the iron required for erythropoiesis by recycling iron from senescent erythrocytes (Weiss, 2002). Iron recycling by macrophages represents the largest pathway of intracellular iron export into the blood system (Ganz & Nemeth, 2006). Ferroportin-1 (FPN1/MTP1/SLC40A1) is a transmembrane transporter protein that plays a key role in the export of iron (Abboud & Haile, 2000; Donovan et al., 2000; McKie et al., 2000). Several mutations in the human FPN1 gene were unequivocally associated with iron accumulation within macrophages, suggesting a critical role of FPN1 in the release of iron from macrophages. Some divalent metals, such as iron and copper, have been shown to affect the level of FPN1 gene expression in vivo and in vitro (Chung et al., 2004; Knutson et al., 2003). It has not yet been determined whether environmentally toxic metals like cadmium can influence FPN1 expression. In the present study, we examined the effects of cadmium on the regulation of FPN1 gene expression in J774 macrophage cells.
J774 mouse macrophage cells were cultured in α-minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum and antibiotics. Before treatments, cells were seeded in a 6-well plate and grown to 60~80% confluence. Stock solutions of CdCl2, actinomycin D, and N-acetylcystein (NAC) were prepared in PBS (pH 7.4) and sterilized by filtration with 0.2 µm membrane.
Total RNA was isolated from the treated cell with Trizol® reagent (Invitrogen, USA). Reverse transcription was carried out with l µg RNA samples by using Reverse transcription master premix (ELPIS biotech, Korea) at 42℃ for 60 min followed by inactivation of reverse transcriptase at 94℃ for 5 min. RT-PCR was performed on MJ mini gradient thermal cycler (Bio-Rad, USA). RT-PCR conditions were 95℃ for 30s, 65℃ for 30s, 72℃ for 1 min with a final extension at 72℃ for 10 min. The primer sets were FPN1 (forward) TTGCAGGAGTCATTGCTGCTA, (reverse) TGGAGTTCTGCACACCATTGAT; β-actin (forward) CTGGCACCACACCTTCTA, (reverse) GGGCACAGTTGGGTGAC (Xenotech, Korea). PCR products were separated on a 2% agarose gel containing ethidium bromide and bands were analyzed with Gel Doc XR system (Bio-Rad, USA) and Quantity on 1-D analysis software (Bio-Rad, USA). For each experiments, the relative abundance value obtained by densitometric analysis was normalized to the value derived from the control sequence (β-actin) in the corresponding sample.
FPN1 promoter/luciferase reporter gene construct (FPN1-Luc, a gift from Dr. Haile, University of Texas at San Antonio, USA) was used for this experiment. The FPN1-Luc plasmid consists of ~2.6 kb of FPN1 5' promoter region, all of the 5'-untranslated region (5'-UTR) including IRE, and firefly luciferase coding sequence. Empty vector that contains firefly luciferase coding sequence only was used as a negative control. For transient transfection, HeLa cells were seeded at 2.7 × 105 cells in a 6-well plate and grown to ~60% confluence. FPN1-Luc plasmid was transiently transfected into HeLa cells using PolyFect® transfection reagent (QIAGEN, USA) according to manufacturer's manual. At 12-h after transfection, cells were washed and divided, and then treated with various concentrations of CdCl2 for 8 h. After metal treatments, cell lysates were prepared using 1× lysis buffer. Luciferase activity was measured with a luminometer using a luciferase reporter assay system (Promega, USA).
After cadmium treatment, cells were washed three times with PBS and lysed in a lysis buffer containing 4× Tris/SDS (pH 6.8), 5% β-mercaptoethanol, and protease inhibitor cocktails. After DNA shearing, protein concentrations of lysates were determined by RC DC Protein assay (Bio-Rad, USA). Fifteen micrograms of cell lysates were separated by SDS/polyacrylamide gel electrophoresis, and transferred to PVDF membranes. The membranes were stained with Ponceau-S solution to visualize and confirm that equal amounts of protein were transferred among wells. After rinsing Ponceau-S solution, the membranes were blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.01% Tween 20 for at least 1~2 hour. Blocked membrane was incubated with affinity purified anti-FPN1 antibody (1:100) overnight at 4℃. The membrane was then washed with TBS-T for 30 min and incubated with secondary anti-rabbit IgG peroxidase-linked antibody (Amersham, USA, 1:5,000) in 2.5% skim milk/TBS-T for at least 1 hour at room temperature. Specific bands were visualized with an enhanced chemiluminescence system (ELPIS biotech, Korea). Intensities of the bands were quantified using a Gel Doc XR system (Bio-Rad, USA) equipped with Quantity one 1-D analysis software (Bio-Rad, USA).
Intracellular ROS was detected by the oxidation-sensitive fluorescent probe dye, 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA) (Invitrogen Molecular Probes, OR, USA). This dye is a stable nonpolar compound which diffuses readily into the cells. Intracellular ROS in the presence of peroxidase changes this dye to the highly fluorescent compound DCF. Thus, the fluorescent intensity is proportional to the amount of peroxides which are produced by the cells. Following exposure to cadmium for 2 hours, the cells were collected and washed with ice-cold PBS. We then added 1 ml of PBS containing 10 µM DCFH-DA, and incubated the cells for 30 min at 37℃. The fluorescence emission from DCF was analyzed via FACSscan flow cytometry (Becton-Dickinson, CA, USA).
The concentration of glutathione (GSH and GSSG) in cellular lysates was determined according to the methods of Rahman et al. (2006). Briefly, washed cells were lysed by repeatedly freezing and thawing using lysis buffer containing 0.6% (w/v) sulfosalicylic acid, 0.15 (v/v) Triton X-100, 5 mM EDTA in 0.1M potassium phosphate buffer, pH 7.5. The supernatant was collected after centrifugation and incubated with 0.2 mg/ml dithiobisnitrobenzoic acid (DTNB) and 1.67 U/ml glutathione reductase in phosphate buffer-EDTA for 30 s, then 0.2 mg/ml β-NADPH was added and the rate of DTNB reduction was spectrophotometrically measured at 405 nm. GSH content was calculated using a standard curve, and expressed as nmol/mg protein. For GSSG measurements, the cell supernatants were first mixed with 2-vinylpyridine and incubated at room temperature for 1 h. After incubation, triethanolamine was added and GSH assay were repeated.
All experiments were repeated three or four times on separate occasions. Statistical analysis was performed by using the SAS systems (ver. 9.1). Differences between groups were tested by ANOVA followed by Duncan's multiple comparison test. All data were expressed as mean ± SD. P values less than 0.05 were considered significant.
To determine cadmium treatment conditions that are non-toxic to J774 cells, we first evaluated the effects of cadmium treatment at various doses and time on the viability of macrophage cells in culture using the MTT assay. J774 macrophage cells (5×104/well, 96 well-plates) were treated with CdCl2 at indicated concentrations for 8, 12 or 20 hours, and incubated with MTT reagent for 4 hours. As shown in Fig. 1, there was a significant loss of viability when the concentrations of CdCl2 was equal to or higher than 50 µM, where the number of viable cells was 60% and 46% of the control after 12 and 20 hours of exposure, respectively. 100 µM CdCl2 further decreased the viable cell numbers to 37% and 27% of the controls after 12- and 20- hour treatment, respectively. On the other hand, incubation of J774 cells with CdCl2 at concentrations of 5 to 25 µM for 8 hours had no significant effect on cell viability. Accordingly, cells were treated with CdCl2 at a concentration lower than 25 µM for shorter than 8 hours in all subsequent experiments to ensure the treatment was noncytotoxic.
To examine whether cadmium affects FPN1 mRNA levels, J774 cells were incubated with different concentrations of CdCl2 (5~25 µM) for 8 hours and total RNAs were extracted for RT-PCR analyses. The results of this analysis showed that cadmium induced FPN1 mRNA expression in a dose-dependent manner. As shown in Fig. 2, the induction of FPN1 mRNA was evident at 5 µM and increased linearly upon incubation with concentrations up to 25 µM. Quantification of FPN1 mRNA levels normalized to β-actin mRNAs levels revealed a 1.5-, 2.5-, and 3- fold increase by 5, 10, and 25 µM CdCl2, respectively. To examine the time required for cadmium-induced FPN1 mRNA expression, cells were treated with 25 µM CdCl2 for 0, 1, 2, 4 or 8 hours. In these experiments, we found that FPN1 was readily induced by cadmium as early as 4 hours and increased further after 8 hours (Fig. 3).
We further examined whether the elevated FPN1 mRNA levels by cadmium treatment were associated with an increase in FPN1 protein levels by Western blot analyses. Basal levels of FPN1 protein in untreated J774 cells were very low, but when cells were treated with 5~25 µM CdCl2 for 8 hours, FPN1 protein levels were greatly increased in a dose-dependent manner (Fig. 4).
To investigate the molecular basis for the induced expression of FPN1 by cadmium, cells were treated with 25 µM CdCl2 in the presence or absence of 0.5 µg/ml actinomycin D for 4 hours. Actinomycin D forms a stable complex with double-stranded DNA, thus inhibiting DNA-primed RNA synthesis, i.e. gene transcription. The results of these experiments showed that actinomycin D treatment alone did not change the steady-state level of FPN1 mRNA in J774 cells. However, increased FPN1 gene expression by cadmium treatment was almost completely blocked when cells were co-treated with actinomycin D (Fig. 5). These results indicate that cadmium up-regulates FPN1 gene expression in a transcription-dependent manner.
To further examine the cadmium-dependent regulation of FPN1 gene transcription, reporter assays using the luciferase expression system were performed. A reporter construct that contained the full-length FPN1 promoter region and luciferase gene were transiently transfected into HeLa cells. 24 hours after transfection, cells were treated with 0 to 25 µM CdCl2 for 8 hours and the luciferase activity of cell lysates was measured using a luminometer. Fig. 6 shows that cadmium treatment greatly increased luciferase activity in a dose-dependent manner, suggesting that cadmium loading into macrophage cells stimulates the promoter activity of the FPN1 gene.
To investigate the possibility that cadmium-induced oxidative stress may play a role in the up-regulation of FPN1 gene expression, we first examined whether intracellular ROS production was increased in cadmium loaded J774 macrophage cells. The ROS production in J774 cells was determined using H2DCFDA fluorescence. As shown in Fig. 7, cadmium treatment significantly increased ROS levels by more than 2-fold compared to the untreated controls (P < 0.05).
We then examined the effects of antioxidant NAC treatment on cadmium-induced FPN1 mRNA expression. J774 cells were pretreated in the presence or absence of 1 mM NAC for 30 min and then further treated with or without 25 µM CdCl2 for 8 hours. Results showed that cadmium treatment alone decreased the intracellular GSH/GSSH ratio to less than half of that found in the untreated controls. When cells were co-treated with NAC, the intracellular GSH/GSSG ratio was recovered to the levels of the untreated controls (Fig. 8A). Furthermore, increased FPN1 mRNA expression by cadmium treatment was attenuated when cells were co-treated with NAC (Fig. 8B, C).
Cadmium intoxication has been associated with the dysregulation of iron homeostasis, which suggests that the expression of iron regulatory proteins can be affected by cadmium stress. In the present study, we clearly demonstrated that cadmium loading into macrophage cells significantly increased FPN1 mRNA levels in a dose- and time-dependent manner. Furthermore, cadmium-induced FPN1 mRNA expression was associated with increased levels of FPN1 protein in macrophages. FPN1 is a key iron exporter in macrophages and iron release from macrophages plays an important role in maintaining iron homeostasis (Aydemir et al., 2009). Therefore, it is possible that elevated concentrations of cadmium result in excess iron release from macrophage cells and disrupt iron homeostasis in the body. In addition, the potent positive effects of cadmium on FPN1 levels suggest that cadmium may also be a transport substrate for export by FPN1. Likewise, DMT1, an iron importer, was shown to be capable of transporting various divalent metals including cadmium (Gunshin et al., 1997). The increased levels of FPN1 in macrophages could result in the acceleration of cadmium export into the circulating system, which would potentiate its toxicity.
In the present study, we showed that cadmium-mediated FPN1 mRNA induction in J774 cells was completely blocked when cells were co-treated with a transcription inhibitor actinomycin D. Moreover, the transactivational activity of the FPN1 promoter was directly stimulated by cadmium loading. Taken together, these results demonstrate that cadmium loading into macrophages up-regulates FPN1 gene expression in a transcription-dependent manner. Cadmium has been shown to regulate the transcription of genes that encode a variety of different types of proteins, including metallothionein, heat shock protein, tumor necrosis factor, heme oxygenase-1, etc. (Bakshi et al., 2008; Lee et al., 2002; Yang et al., 2007). Cadmium can act as a strong ligand for a transcription factor called MTF (metal response element (MRE)-binding transcription factor-1), which binds to MRE in the promoter region of cadmium-regulated genes. However, it is not clear whether the FPN1 gene contains functional MREs in its promoter region. In addition, previous studies have shown that zinc, another potent activator of MTF, didn't enhance FPN1 mRNA levels in macrophages (Chung et al., 2004; Park & Chung, 2008). Therefore, it is likely that regulation of FPN1 gene expression by cadmium is independent of the MTF-MRE system.
We found that cadmium loading into macrophage cells significantly increased intracellular ROS production. Similar to our observation, treatment of microglia cells, resident macrophages of the brain, with cadmium was shown to induce oxidative stress as assessed by increased DMPO-trapped electron spin resonance (ESR) signals and intracellular ROS levels (Yang et al., 2007). The ability of cadmium to induce ROS formation has also been described by other studies in different cell types (Koizumi et al., 1996; Thevenod et al., 2000). Furthermore, it was reported that in vivo cadmium exposure also led to excessive production of ROS in mouse peritoneal macrophages (Ramirez & Gimenez, 2003). Since cadmium is a divalent ion at physiological pH, it is unlikely that the metal directly generates ROS. In the present study, we demonstrated that cadmium treatment significantly decreased the GSH/GSSG ratio. Our results are consistent with previous studies, which reported that cadmium exposure resulted in GSH depletion and increased oxidative stress in brain cells and murine thymocyts (Im et al., 2006; Lopez et al., 2006; Pari & Murugavel, 2007; Pathak & Khandelwal, 2007). GSH is the predominant antioxidant in the cytoplasm of cells and can scavenge ROS. Therefore, it appears that cadmium indirectly increases intracellular ROS levels through the depletion of antioxidant molecules.
To further investigate whether the cadmium-induced oxidative stress is associated with changes in FPN1 mRNA, we treated J774 cells with the antioxidant NAC. NAC is an important inducer of intracellular glutathione and has been shown to be a ROS scavenger (Gaubin et al., 2000; Shaikh et al., 1999). In the present study, we found that the intracellular GSH/GSSG ratio was increased when J774 cells were co-treated with cadmium and NAC, compared to when cells were treated with cadmium alone. More importantly, cadmium-induced FPN1 mRNA expression was markedly attenuated when cells were co-treated with NAC. The results of the present study strongly indicate that cadmium enhances FPN1 gene transcription via increased production of ROS. Several proteins that are related to iron metabolism are shown to be redox sensitive. For example, H2O2 treatment significantly increased the levels of the transferrin receptor (TfR), the major iron uptake protein (Sureda et al., 2005). Also, it is well known that the expression of ferritin, the major iron storage protein, can be modulated in response to a battery of different oxidative stressors such as H2O2, tert-butylhydroquinone, hemin, and UV-irradiation (Hintze & Theil, 2006; Iwasaki et al., 2006; Tsuji, 2005; Tsuji et al., 2000). Both TfR and ferritin transcripts contain an iron responsive element (IRE), where iron regulatory proteins (IRP1 and IRP2) can specifically bind to. IRPs are proposed as sensors of ROS, since their activities are readily changed by ROS. The mRNA of FPN1 also contains putative IREs in its 5'-untranslated region. Thus, further studies are needed to determine whether the IRP/IRE system is involved in cadmium-induced FPN1 expression.
In summary, the results of this study clearly demonstrate that cadmium significantly increased the expression of iron exporter FPN1 in macrophages, suggesting another important interaction between cadmium and iron metabolism. The interaction of cadmium and iron metabolism has also been shown in other cell types such as enterocytes (Ryu et al., 2004). Future studies are warranted to investigate the physiological consequences of cadmium-induced FPN1 expression on iron homeostasis and cadmium toxicity.
Figures and Tables
Fig. 1
Effects of cadmium treatment on the viability of J774 macrophage cells. J774 cells were treated with CdCl2 with indicated concentrations and time. The cell viability was measured by MTT assay as described in Methods and Materials. Data shown are mean ± SD (n=6). *P < 0.05 vs. untreated controls

Fig. 2
Effects of cadmium treatment with various concentrations on FPN1 mRNA levels in J774 cells. J774 cells were incubated with the indicated concentrations of CdCl2 for 8 h. Total RNA were prepared and analyzed for FPN1 or β-actin mRNA by RT-PCR analyses. Data shown are representative agarose gels (upper panel) or means ± SD of three independent experiments (lower panel). *P < 0.05 vs. untreated controls

Fig. 3
Effects of cadmium treatment with various incubation time on FPN1 mRNA levels in J774 cells. J774 cells were incubated with 25 µM CdCl2 for the indicated time. Total RNA were prepared and analyzed for FPN1 or β-actin mRNA by RT-PCR analyses. Data shown are representative agarose gels (upper panel) or means ± SD of three independent experiments (lower panel). *P < 0.05 vs. untreated controls

Fig. 4
Effects of cadmium treatment on FPN1 protein levels in J774 cells. J774 cells were incubated with 0, 5, 10, or 25 µM CdSO4 for 8 h. Whole cell lysates were prepared and analyzed for FPN1 protein by Western blot. Upper panel: representative blot, Lower panel: band density detected by chemiluminescence was quantified (QuantityOne imaging software, Bio-Rad). Values are mean ± SD of three independent experiments. *P < 0.05 vs. untreated controls
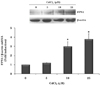
Fig. 5
Effects of cadmium and actinomycin D on FPN1 mRNA levels in J774 cells. J774 cells were treated with 25 µM CdCl2 for 4 h in the presence or absence of 0.5 µg/ml actinomycin D. Total RNAs were prepared and analyzed for FPN1 or β-actin mRNA by RT-PCR. Upper panel: representative agarose gels, Lower panel: band intensity detected by ethidium bromide-staining was quantified (QuantityOne imaging software, Bio-Rad). Values are mean ± SD of three independent experiments. *P < 0.05 vs. untreated controls

Fig. 6
Effects of cadmium treatment on the luciferase activity driven by FPN1-promoter. HeLa cells were transiently transfected with FPN1 promoter/luciferase reporter constructs for 12 h. Cells were treated with 0, 5, 10, or 25 µM CdSO4 for another 8 h. Luciferase activity in cell lysates was then determined. Values are presented as mean ± SD of three independent experiments. *P < 0.05 vs. untreated controls

Fig. 7
Effects of cadmium treatment in J774 cells on the reactive oxygen species (ROS) generation. J774 cells were treated with 25 µM cadmium and intracellular ROS generation was detected by using 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA). Values are presented as mean ± SD of three independent experiments. *P < 0.05 vs. untreated controls

Fig. 8
Effects of cadmium and N-acetylcystein (NAC) treatment on FPN1 mRNA levels in J774 cells. J774 cells were pretreated in the presence or absence of 1 mM NAC for 30 min, and then further treated with or without 25 µM CdCl2. Total RNA were prepared and analyzed for FPN1 or β-actin mRNA by RT-PCR analyses. Data shown are representative agarose gels (A) or means ± SD of three independent experiments (B). Intracellular GSH/GSSG ratio was measured as described in Methods and Materials (C). *P < 0.05 vs. untreated controls

References
1. Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000. 275:19906–19912.

2. Agency for Toxic Substance and Disease Registry. Toxicological Profile for Cadmium. 2003. Atlanta, GA. USA: U.S. Department of Health and Humans Services, Public Health Service, Centers for Disease Control.
3. Aydemir F, Jenkitkasemwong S, Gulec S, Knutson MD. Iron loading increases ferroportin heterogeneous nuclear RNA and mRNA levels in murine J774 macrophages. J Nutr. 2009. 139:434–438.

4. Bakshi S, Zhang X, Godoy-Tundidor S, Cheng RY, Sartor MA, Medvedovic M, Ho SM. Transcriptome analyses in normal prostate epithelial cells exposed to low-dose cadmium: Oncogenic and immunomodulations involving the action of tumor necrosis factor. Environ Health Perspect. 2008. 116:769–776.

5. Bressler JP, Olivi L, Cheong JH, Kim Y, Bannona D. Divalent metal transporter 1 in lead and cadmium transport. Ann N Y Acad Sci. 2004. 1012:142–152.

6. Chung J, Haile DJ, Wessling-Resnick M. Copper-induced ferroportin-1 expression in J774 macrophages is associated with increased iron efflux. Proc Natl Acad Sci U S A. 2004. 101:2700–2705.

7. Djukic-Cosic D, Curcic Jovanovic M, Plamenac Bulat Z, Ninkovic M, Malicevic Z, Matovic V. Relation between lipid peroxidation and iron concentration in mouse liver after acute and subacute cadmium intoxication. J Trace Elem Med Biol. 2008. 22:66–72.

8. Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, Law TC, Brugnara C, Lux SE, Pinkus GS, Pinkus JL, Kingsley PD, Palis J, Fleming MD, Andrews NC, Zon LI. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000. 403:776–781.

9. Ganz T, Nemeth E. Regulation of iron acquisition and iron distribution in mammals. Biochim Biophys Acta. 2006. 1763:690–699.

10. Gaubin Y, Vaissade F, Croute F, Beau B, Soleilhavoup J, Murat J. Implication of free radicals and glutathione in the mechanism of cadmium-induced expression of stress proteins in the A549 human lung cell-line. Biochim Biophys Acta. 2000. 1495:4–13.

11. Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997. 388:482–488.

12. Hintze KJ, Theil EC. Cellular regulation and molecular interactions of the ferritins. Cell Mol Life Sci. 2006. 63:591–600.

14. Im JY, Paik SG, Han PL. Cadmium-induced astroglial death proceeds via glutathione depletion. J Neurosci Res. 2006. 83:301–308.

15. International Agency for Research on Cancer. Berrylium, cadmium, mercury and exposures in the glass manufacturing industry. International Agency for Research on Cancer Monographs on the Evaluation of Carcinogenic Risks to Humans. 1993. 58. Lyon. France: IARC Scientific Publications;119–237.
16. Iwasaki K, Mackenzie EL, Hailemariam K, Sakamoto K, Tsuji Y. Hemin-mediated regulation of an antioxidant-responsive element of the human ferritin H gene and role of ref-1 duringerythroid differentiation of K562 cells. Mol Cell Biol. 2006. 26:2845–2856.

18. Järup L, Berglund M, Elinder CG, Nordberg G, Vahter M. Health effects of cadmium exposure--a review of the literature and a risk estimate. Scand J Work Environ Health. 1998. 24:Suppl 1. 1–51.
19. Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003. 102:4191–4197.

20. Koizumi T, Shirakura H, Kumagai H, Tatsumoto H, Suzuki KT. Mechanism of cadmium-induced cytotoxicity in rat hepatocytes: Cadmium-induced active oxygen-related permeability changes of the plasma membrane. Toxicology. 1996. 114:125–134.

21. Lee MJ, Nishio H, Ayaki H, Yamamoto M, Sumino K. Upregulation of stress response mRNAs in COS-7 cells exposed to cadmium. Toxicology. 2002. 174:109–117.

22. Lopez E, Arce C, Oset-Gasque MJ, Canadas S, Gonzalez MP. Cadmium induces reactive oxygen species generation and lipid peroxidation in cortical neurons in culture. Free Radic Biol Med. 2006. 40:940–951.

23. McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000. 5:299–309.

24. Pari L, Murugavel P. Diallyl tetrasulfide improves cadmium induced alterations of acetylcholinesterase, ATPases and oxidative stress in brain of rats. Toxicology. 2007. 234:44–50.

25. Park BY, Chung J. Effects of various metal ions on the gene expression of iron exporter ferroportin-1 in J774 macrophages. Nutrition Research and Practice. 2008. 2:317–321.

26. Pathak N, Khandelwal S. Role of oxidative stress and apoptosis in cadmium induced thymic atrophy and splenomegaly in mice. Toxicol Lett. 2007. 169:95–108.

27. Rahman I, Kode A, Biswas SK. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006. 1:3159–3165.

28. Ramirez DC, Gimenez MS. Induction of redox changes, inducible nitric oxide synthase and cyclooxygenase-2 by chronic cadmium exposure in mouse peritoneal macrophages. Toxicol Lett. 2003. 145:121–132.

29. Ryu DY, Lee SJ, Park DW, Choi BS, Klaassen CD, Park JD. Dietary iron regulates intestinal cadmium absorption through iron transporters in rats. Toxicol Lett. 2004. 152:19–25.

30. Satarug S, Baker JR, Urbenjapol S, Haswell-Elkins M, Reilly PE, Williams DJ, Moore MR. A global perspective on cadmium pollution and toxicity in non-occupationally exposed population. Toxicol Lett. 2003. 137:65–83.

31. Schafer SG, Schwegler U, Schumann K. Retention of cadmium in cadmium-naive normal and iron-deficient rats as well as in cadmium-induced iron-deficient animals. Ecotoxicol Environ Saf. 1990. 20:71–81.

32. Shaikh ZA, Vu TT, Zaman K. Oxidative stress as a mechanism of chronic cadmium-induced hepatotoxicity and renal toxicity and protection by antioxidants. Toxicol Appl Pharmacol. 1999. 154:256–263.

33. Staessen JA, Buchet JP, Ginucchio G, Lauwerys RR, Lijnen P, Roels H, Fagard R. Public health implications of environmental exposure to cadmium and lead: An overview of epidemiological studies in belgium. working groups. J Cardiovasc Risk. 1996. 3:26–41.

34. Sureda A, Hebling U, Pons A, Mueller S. Extracellular H2O2 and not superoxide determines the compartment-specific activation of transferrin receptor by iron regulatory protein 1. Free Radic Res. 2005. 39:817–824.

35. Thevenod F, Friedmann JM, Katsen AD, Hauser IA. Up-regulation of multidrug resistance P-glycoprotein via nuclear factor-kappaB activation protects kidney proximal tubule cells from cadmium- and reactive oxygen species-induced apoptosis. J Biol Chem. 2000. 275:1887–1896.
36. Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005. 24:7567–7578.

37. Tsuji Y, Ayaki H, Whitman SP, Morrow CS, Torti SV, Torti FM. Coordinate transcriptional and translational regulation offerritin in response to oxidative stress. Mol Cell Biol. 2000. 20:5818–5827.

38. Yang Z, Yang S, Qian SY, Hong JS, Kadiiska MB, Tennant RW, Waalkes MP, Liu J. Cadmium-induced toxicity in rat primary mid-brain neuroglia cultures: Role of oxidative stress from microglia. Toxicol Sci. 2007. 98:488–494.

40. World Health Organization. Chapter 6.3. Cadmium. 2000. Geneva. Switzerland: 1–11.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download