Dear Editor,
B-cell ALL (B-ALL) with double homeobox 4 rearrangement (
DUX4r) is a recently identified molecular subtype characterized by rearrangements involving the
DUX4 transcription factor gene, typically with the immunoglobulin heavy chain (
IGH) or ETS transcription factor ERG (
ERG) locus [
1]. This subtype accounts for 5%–10% of B-ALL cases, predominantly affects adolescents and young adults, and demonstrates favorable outcomes despite poor minimal residual disease (MRD) responses [
2]. Thus, identifying
DUX4r enables risk stratification and tailored treatment. However, detecting
DUX4 mutations is challenging owing to its repetitive, multilocus region within the GC-rich D4Z4 array at subtelomeric 4q35 and 10q26, thereby complicating identification with conventional cytogenetic techniques [
3–
5].
Studies making efforts to detect
DUX4r have proposed
ERG deletion [
4] and immunophenotyping [
6] markers, such as CD371 and CD2, as indicators; however, these fail to identify
DUX4r in some cases [
7]. RNA-sequencing (RNA-seq) is widely used for
DUX4r detection because it helps identify gene fusions and gene expression profiles (GEPs) [
4,
8]. However, fusion-calling algorithms may miss
DUX4r, even when the GEP suggests its presence [
9]. Whole-genome sequencing (WGS) has proven effective in identifying
DUX4r by providing detailed information about breakpoint locations [
3,
8].
Here, we present a case of an 8-year-old girl diagnosed with B-ALL with
IGH::DUX4 rearrangement, highlighting the diagnostic challenges and utility of advanced genomic techniques. She had no known medical history and was referred to Severance Hospital, Seoul, Korea, in August 2023 with severe anemia. This study was approved by the Institutional Review Board of Severance Hospital, Seoul, Korea (4-2024-1223). Informed consent was waived because the study used anonymized data. Upon admission, her complete blood cell count showed a Hb level of 54 g/L, platelet count of 152×10
9/L, and white blood cell count of 8.67×10
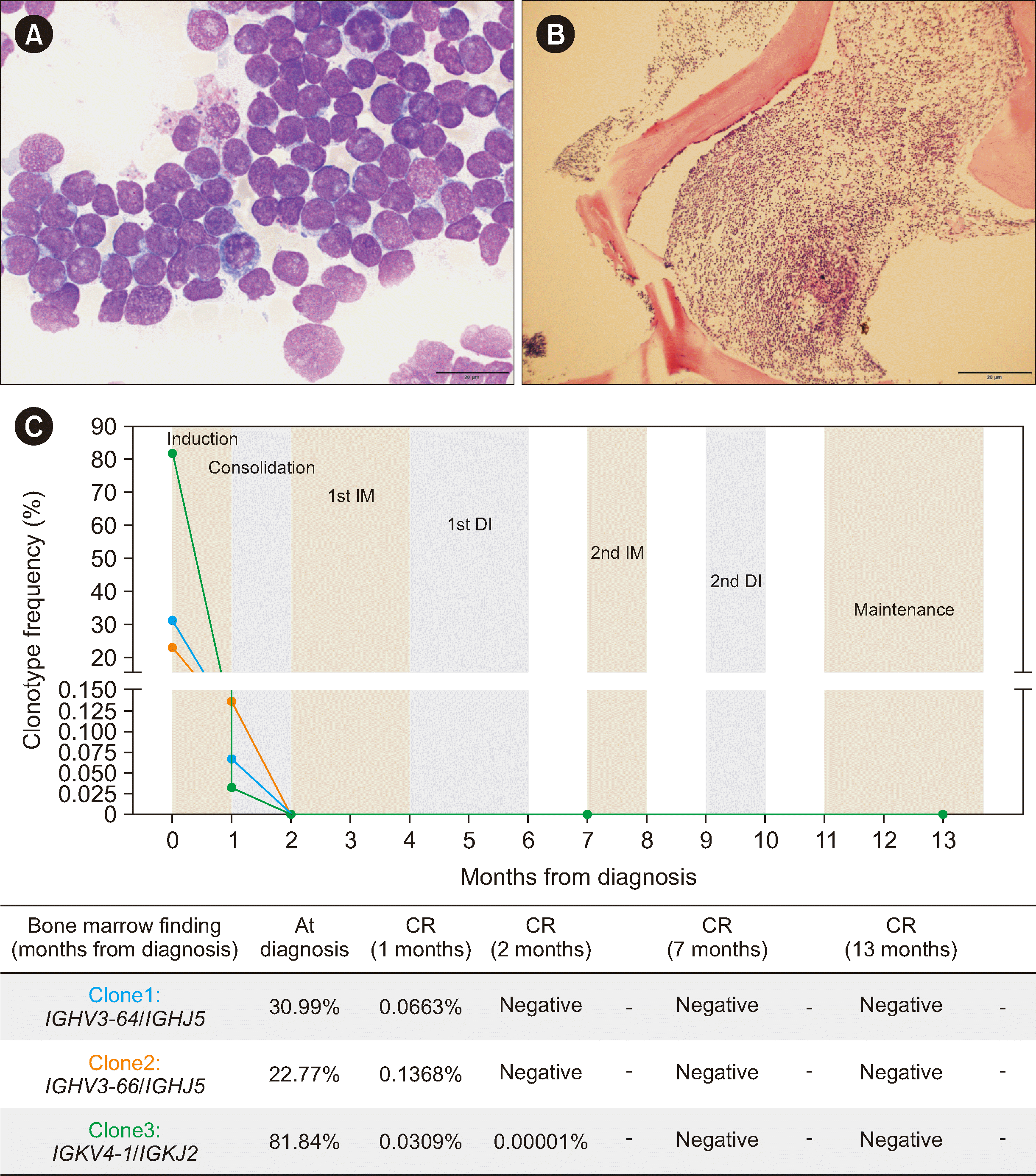
9/L. Leukemic blasts were observed on her peripheral blood smear, and her bone marrow was packed with lymphoblasts as indicated by flow cytometry (
Fig. 1A and
1B). Blasts were positive for CD34, cCD79a, CD19, CD38, HLA-DR, cCD22, nTdT, and CD10. Markers CD371 and CD2, recommended for identifying
DUX4r, were not assessed. Concurrent next-generation sequencing detected three tier 1/2 mutations in
CREBBP: NM_004380.3, c.2302C>T (p.Arg768Ter); c.1610_1611insCATC (p.Thr538IlefsTer34); and c.445C>T (p.Gln149Ter). No intragenic
ERG deletion was detected.
 | Fig. 1
Morphologic and molecular characterization of B-ALL with IGH::DUX4 rearrangement, including minimal residual disease tracking. (A) Bone marrow biopsy (magnification, 1×) revealing a bone marrow packed with blasts. (B) Blasts display a high nuclear-to-cytoplasmic ratio with round nuclei and prominent nucleoli (magnification, 100×). (C) Changes in IGH and IGK clonotypes over time in response to treatment, indicating minimal residual disease. During diagnosis, clonotype frequency was presented as a percentage of total lymphocytes. At follow-up, it was adjusted to represent a percentage of total cells using LymphoQuant Internal Controls (Invivoscribe Technologies, San Diego, CA, USA).
Abbreviations: CR, complete remission; IM, interim maintenance; DI, delayed intensification.

|
RNA-seq was performed using a TruSeq Stranded mRNA kit (Illumina, San Diego, CA, USA) on a NextSeq 550Dx (Illumina). While the fusion-calling algorithms Arriba and STAR did not detect
DUX4r or any other fusion, the GEP suggested the possibility of
DUX4r (
Fig. 2A). To confirm
DUX4r, we performed long-read WGS using the Sequel II system (Pacific Biosciences, Menlo Park, CA, USA), focusing on the region between
IGHJ4 and
IGHD2-15, based on the
IGH breakpoint observed in NALM-6 cells known to carry the
IGH::
DUX4 translocation (
Fig. 2B) [
8]. The sequencing results confirmed an
IGH::
DUX4 rearrangement, leading to a definitive diagnosis of B-ALL with
IGH::
DUX4 rearrangement. Expression profiles and
IGH breakpoints in
IGH::
DUX4- positive B-ALL samples reportedly closely resemble those of NALM-6 [
3].
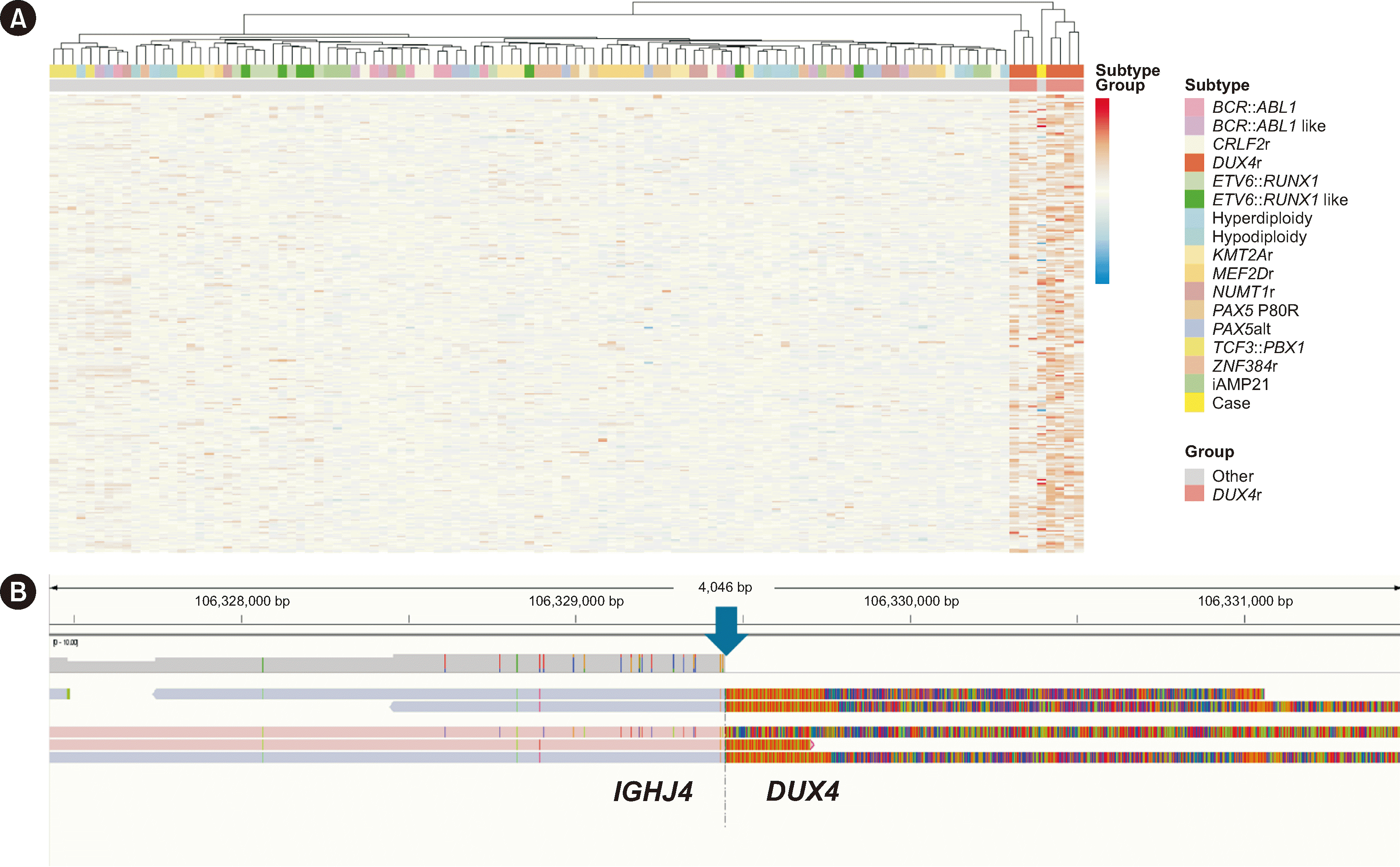 | Fig. 2 Transcriptomic and genomic characterization of B-ALL with IGH::DUX4 rearrangement. (A) Heatmap of differentially expressed genes (DEGs) identified by edgeR. Data for this analysis were obtained from the Genomics Platform (St. Jude Cloud, https://www.stjude.cloud/) and included 16 ALL subtypes, each represented by seven randomly sampled cases. Our case data were processed into feature counts according to the St. Jude RNA-seq expression classification workflow (https://platform.stjude.cloud/workflows/rnaseq-expression-classification) and are represented as “Case” within the “Other” group, which comprises 15 ALL subtypes excluding DUX4r. Gene expression profiles were derived from a matrix of DEGs identified through edgeR analysis (https://bioconductor.org/packages/release/bioc/html/edgeR.html), with the DUX4r group compared with the “Other” group as a reference. DEGs were selected based on significantly higher or lower expression in the DUX4r group, with thresholds of |log2FC| >1 and an adjusted P<0.0001. Our case (“Case”) clustered within the DUX4r group, suggesting alignment in the expression profile with the DUX4r subtype. (B) Identification of the DUX4r breakpoint (blue arrow) within the IGHJ4 region on chromosome 14. Illustrated is an enlarged view of the DUX4r locus on chromosome 14, GRCh37 reference genome, between positions 106,324,391 and 106,334,508, viewed using Integrative Genomics Viewer (Broad Institute, Cambridge, MA, USA). Previous studies have identified that the breakpoint characteristic of the DUX4r subtype of ALL often occurs within the region between IGHJ4 and IGHD2-15 [3, 8]. Guided by these findings, we examined the genomic location of IGHJ4 (chr14: 106,322,261 – 106,332,378) and detected the presence of DUX4r at this locus.
|
The patient underwent standard induction chemotherapy, and MRD was tested on the three clonotypes identified at diagnosis using the Lymphotrack
IGH FR1 and
IGK assays (Invivoscribe Technologies, San Diego, CA, USA). The patient showed complete remission with an MRD >0.1% after induction chemotherapy and <0.01% after consolidation chemotherapy (
Fig. 1C). Currently, the patient is undergoing maintenance chemotherapy and remains in molecular complete remission.
Given the genetic profile of
DUX4r-positive B-ALL, somatic mutations commonly associated with this subtype include lymphoid transcription factor genes such as
IKZF1 and
PAX5 [
4]. However, in this case, only a
CREBBP mutation was identified. Although
CREBBP mutations are often associated with relapsed ALL and treatment resistance [
10], their prognostic implications in
DUX4r cases remain unclear and require further investigation.
RNA-seq, WGS, and the use of CD371 and CD2 markers each have distinct strengths and limitations in
DUX4r detection. RNA-seq is cost-effective, helps identify GEPs, and provides co-occurring mutation insights, but it cannot detect structural rearrangements such as
DUX4r. WGS is highly sensitive and specific for structural variations such as
IGH::DUX4 but is resource-intensive. CD371 and CD2 markers aid in large-scale screening but lack the specificity achievable using RNA-seq and WGS. Currently, RNA-seq offers the best balance of cost-effectiveness and comprehensive genomic insights for clinical applications [
11,
12].
In conclusion, while fusion-calling algorithms failed to detect DUX4r, GEP analysis and confirmatory WGS provided an accurate diagnosis. This case emphasizes the importance of comprehensive genomic profiling, including RNA-seq and WGS, in accurately diagnosing complex rearrangements such as IGH::DUX4 in B-ALL.






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download