Abstract
Objective
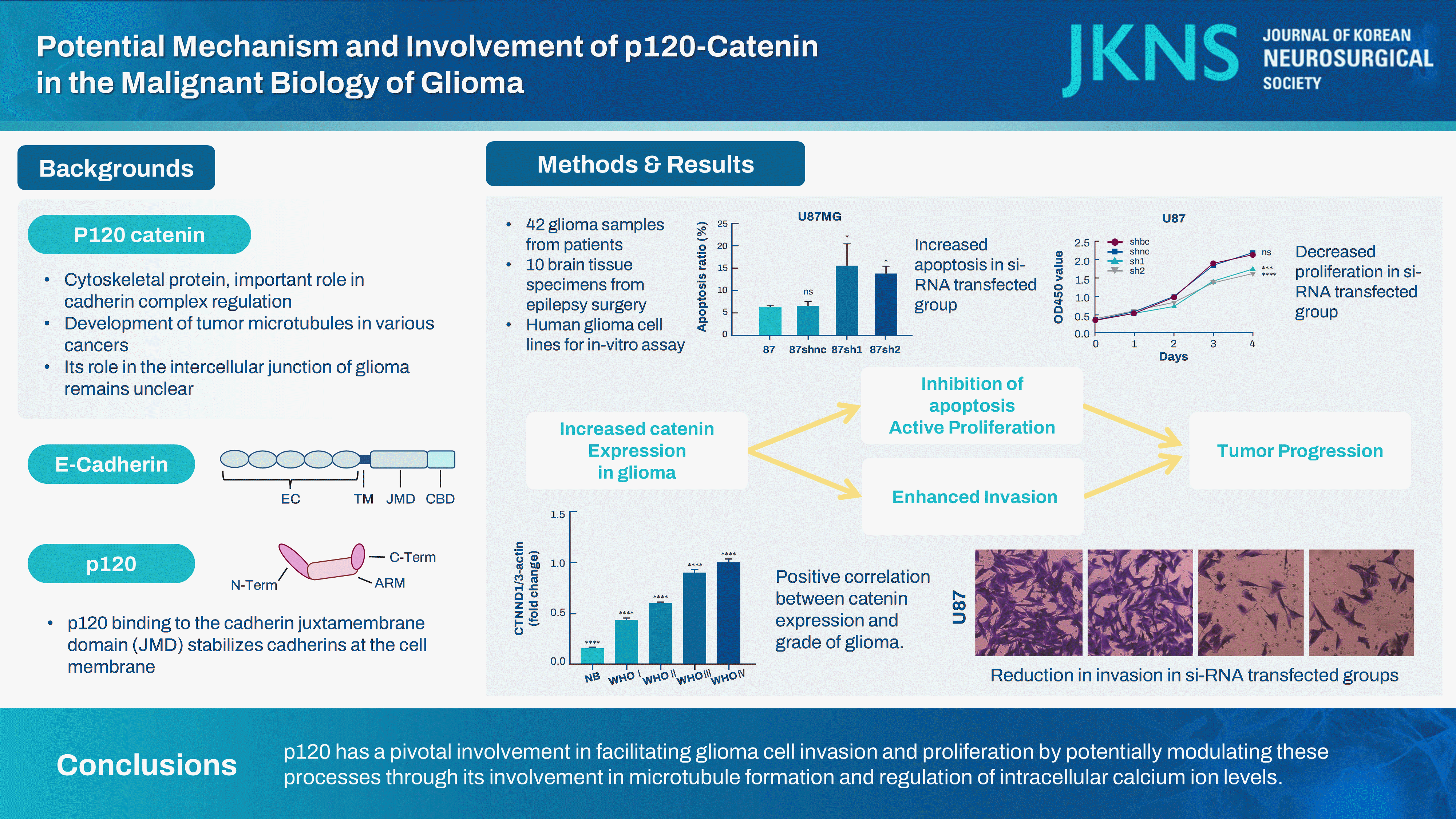
This study analyzed the influence of p120-catenin (catenin [cadherin-associated protein], delta 1 [CTNND1]) on the malignant characteristics of glioma and elucidated the potential underlying mechanism.
Methods
The p120 expression level was assessed in the brain tissues of 42 glioma patients and 10 patients with epilepsy by using the immunohistochemical method. Meanwhile, quantitative polymerase chain reaction (QT-PCR) technology was employed to assess the expression of p120 in the brain tissues of 71 glioma patients and 13 epilepsy patients. LN229, U251, and U87 glioma cells were used for in vitro analysis and categorized into four treatment groups : siRNA-blank control (BC) group (no RNA sequence was transfected), siRNA-negative control (NC) group (transfected control RNA sequences with no effect), and siRNA-1 and siRNA-2 groups (two p120-specific interfering RNA transfection). p120 expression in these treatment groups was quantified by western blotting assay. The migratory and invasive capabilities of glioma cells were studied by wound healing assay and Transwell invasion assay, respectively, under different treatment conditions. MTT (3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide) assay and cell cycle and apoptosis assay were used to determine glioma cell proliferation and apoptosis, respectively. Enzymelabeled assay was performed to measure intracellular calcium ion concentration. Immunofluorescence assay was performed for determining microtubule formation and glioma cell distribution.
Results
Brain tissues of the glioma group exhibited a remarkable increase in the p120 expression level as compared to brain tissues of the nontumor group (p<0.05). Furthermore, a strong positive correlation was noted between the malignancy degree in glioma brain tissues and p120 expression in Western blotting (r=0.906, p<0.0001) and QT-PCR (F=830.6, p<0.01). Compared to the BC and NC groups, the siRNA transfection groups showed a significant suppression in p120 expression in glioma cells (p<0.05), with a marked attenuation in the invasive, migratory, and proliferative capabilities of glioma cells as well as an increase in apoptotic potential (p<0.05). Enzyme-labeled assay showed a remarkable increase in calcium concentration in glioma cells after siRNA treatment. Immunofluorescence assay revealed that the microtubule formation ability of glioma cells reduced after siRNA treatment.
Microtubule is a critical component of the cytoskeleton. Because of their dynamic properties, which include continuous polymerization and depolymerization, microtubules play a pivotal role in diverse physiological cell processes [3,6,21]. Glioma’s heterogeneity significantly influences the progression and recurrence of this prevalent malignant tumor within the brain. According to previous research, the heterogeneous progression of glioma is significantly influenced by the tumor microenvironment (TME) [9,38]. Recently, gliomas have been shown to contain tumor microtubes (TMs), which resemble neurite-like cell protrusions; these TMs play a crucial role in the TME [15,31]. The majority of membrane nanotubes, including TMs, have a high concentration of actin filaments. By using these intercellular connections, tumors can facilitate the exchange of small molecules and toxic substances, thereby influencing localized cellular homeostasis [4,13,30]. Thus, by employing this approach, peripheral glioma cells establish connections within a vast syncytium, thereby forming a functionally and mechanically cohesive network of tumor cells that may potentially impede therapeutic efficacy. GAP43, a neural tissue-specific protein and one of the main regulatory proteins of TMs [7,14,32], is thought to be exclusively expressed in mature neurons and is closely associated with neural development, axon regeneration, and synaptic processes. Actin cytoskeleton has a critical role in microtubule formation; furthermore, the distal end of GAP43 is involved in regulating microtubule formation.
p120, a cytoskeletal protein, is encoded on chromosome 11q11, and it has an important role in cadherin complex regulation. This protein comprises an N-terminal regulatory domain, a C-terminal tail, and a central domain. According to previous studies, p120-catenin is involved in regulating E-cadherin activity, thus indicating that it can bind to the juxta membrane domain of E-cadherin. This binding effectively enhances E-cadherin abundance, leading to the restoration of innate epithelial cell morphology and stabilization of intercellular adhesion [8,11,36]. Additional functions of p120 include the stabilization of transmembrane cadherin molecules at cell-cell junctions, regulation of cellular barrier-associated actin dynamics, platelet pseudopodia formation, and modulation of cell movement by regulating GTPases such as RhoA and Rac [12]. The p120-catenin protein (catenin [cadherin-associated protein], delta 1 [CTNND1]) is an integral component of the synaptic adhesion molecule complex and participates in ensuring cell adhesion stability and regulating dendritic spine formation. As a regulatory protein upstream of GAP43, p120 contributes to the development of TMs in various cancerous tumors [5,16]. However, its role in the intercellular junction of glioma remains unclear. Therefore, the present study analyzed how p120-catenin is involved in influencing glioma malignant characteristics and elucidated the potential underlying mechanism.
The Ethics Committee of The Institutional Review Board of Tianjin Medical University General Hospital (IRB 2024-YX-099-01) approved this study. Written informed consent was obtained from all participants in advance.
We used the following kits and reagents for this study : p120 (CTNND1) antibodies (Affinity Biosciences, Melbourne, Australia); fluo-4 calcium ion detection kit (Beyotime Biotechnology, Shanghai, China); coraLite 488-conjugated goat anti-mouse IgG (Invitrogen Life Technologies, Carlsba, CA, USA); tubulin mouse monoclonal antibodies (Beyotime Biotechnology); 4% paraformaldehyde (Solarbio, Beijing, China); triton X-100 (Solarbio); LipofectamineTM 2000 (Invitrogen, Carlsba, CA, USA); and 4ʹ,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, USA).
From July 2020 to November 2023, 42 tumor samples were obtained from glioma patients who had undergone neurosurgery. Prior to the surgical procedure, no radiotherapy or chemotherapy was administered to any of the patients. A neuropathologist assessed the glioma cases to provide a pathological diagnosis and grading, in accordance with the World Health Organization (WHO) 2021 guidelines for classifying neoplasms of the nervous system. The specimens comprised six, 10, nine, and 17 specimens of WHO grade diffuse astrocytoma, oligodendroglioma, anaplastic oligodendroglioma, and glioblastoma, respectively. For the control group, 10 brain tissue specimens were obtained from epilepsy patients with craniotomy; these specimens were histologically confirmed as non-neoplastic. The tissues were utilized for immunohistochemical investigations. Quantitative polymerase chain reaction (QT-PCR) was employed in 71 cases of glioma with grades ranging from II to IV and also in 13 cases involving epileptic brain. This included 23 cases of grade II, 25 cases of grade III, and 25 cases of grade IV.
Tissue sample collection was based on the protocols validated by the Institutional Review Board of Tianjin Medical University General Hospital (IRB 2024-YX-099-01). Immediately following surgical excision, the samples were frozen and stored in liquid nitrogen for further studies.
LN229, U87, and U251 human glioma cell lines were supplied by the Cell Bank of the Chinese Academy of Sciences. We cultured these cell lines at 37°C in DMEM (Dulbecco’s modification of Eagle’s medium Dulbecco) with 10% fetal bovine serum (FBS) (Solarbio) in a 5% CO2 environment and subjected them to routine passage every 2–3 days.
For immunohistochemical staining, the tissue sections were incubated at 4°C overnight with CTNND1 (p120) antibodies (diluted 1 : 100; Affinity Biosciences, Princeton, NJ, USA). Subsequently, the sections were incubated with 1 : 100 diluted biotinylated secondary antibodies at room temperature (RT) for 30 minutes; this was followed by incubation with ABC peroxidase for another 30 minutes. After a thorough wash with Tris buffer, the sections were stained with 3,3ʹ-diaminobenzene (dissolved in Tris buffer with H2O2 [0.03%] at 30 mg/100 mL concentration) for 5 minutes. The sections were washed with water and counterstained with hematoxylin.
Knockdown of p120-ctn in glioma cells was performed using a siRNA oligonucleotide duplex synthesized by Hanbio (Shanghai, China). The siRNA was generated based on the following human p120-ctn gene sequences : 5'-GGCAGAU UGUGGAGACCUATT-3' and 5'-GCAUGAGCGAGGAAGU UUATT-3'. The 5'-UUCUCCGAACGUGUCACGUTT-3' sequence was used as a negative control. These three types of siRNAs were dissolved separately in diethypyrocarbonate water. For the siRNA-1, siRNA-2, and siRNA-NC groups, 5 µL each of the siRNA or the negative control was mixed with LipofectamineTM 2000 transfection reagent (5 µL) in 500 µL of serum-free medium. Furthermore, the blank control group contained only the transfection reagent.
The tissue specimens were dissected into small fragments weighing 50 mg each and subsequently transferred to individual 1.5 mL microcentrifuge tubes. Next, cell lysis buffer (500 µL) was added to the tube. Subsequently, the mixture was repeatedly homogenized on ice for 10–15 cycles by using a mini-mixer and a plastic pestle, with each cycle lasting for 3–4 seconds. The specimens were then centrifuged at 12000 g at 4°C for 15 minutes. Subsequently, the supernatant was transferred carefully to new tubes for further analysis. Next, 50 µg protein with an equivalent amount of 2×sample buffer was heated for 5 minutes at 94°C. After 48 hours of treatment, the whole cell lysates were prepared for imprinting. The cellular components were separated by SDS (dodecyl sulfate, sodium salt)-PAGE (polyacrylamide gel electrophoresis) (0.1 mol/L Tris [pH 6.8], 20% SDS, 0.2% glycerol, and 0.2 mol/l DL-dithiothreitol) and subjected to a boiling process at 94°C for 5 minutes. The proteins were then separated using a gel composed of 10% SDS-polyacrylamide; the separated proteins were further transferred to polyvinylidene fluoride membranes. Following blocking with phosphate buffer solution and 5% skimmed milk powder at 37°C for 1 hour, the membranes were incubated at 4°C overnight with primary antibodies (p120, anti-rabbit, diluted to 1 : 1000), followed by treatment with mouse anti-rabbit secondary antibodies (diluted to 1 : 5000). The membranes were then treated with an improved electrochemiluminescence chemiluminescence reagent (Amersham Pharmacia, Buckinghamshire, UK). Subsequently, the membranes were imaged using an imaging system (Gene Genius, Frederick, MD, USA). The presence of p120 was identified by p120 antibodies. β-Actin was utilized as the reference standard.
The TRIZOL (total RNA extraction) reagent was utilized to induce denaturation in the glioma tissue, followed by subsequent steps including RNA extraction, precipitation, washing, and dissolution. Following this, an assessment of the concentration and integrity of the RNA was conducted for detection purposes. Subsequently, cDNA underwent amplification using RT-PCR. Quantitative PCR was performed under specific conditions : a 30-second pre-denaturation at 95°C, followed by denaturation at 95°C for 5 seconds and annealing/extension at 62°C for 30 seconds over a total of 40 cycles. The forward primer sequence used for CTNND1 was identified as 5'-CTGTGGAGACCTCAGATGATGG-3' with the reverse primer sequence being noted as 5'-TGGCTGTACTGTCCGTGTTG-3'. Additionally, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) functioned as an endogenous control with its respective forward primer being recorded as 5'-AAGATCATCAGCAATGCCTCCT-3' and the reverse primer as being noted as 5'-CATGAGTCCTTCCACGATACCA-3'.
A medium with 10% FBS was used for preparing a cell suspension. Subsequently, individual wells of 96-well plates were seeded with the cells at 1000 to 10000 cells per well density, and each well was filled with 200 µL of the cell suspension. After 72 hours incubation, 5 mg/mL MTT solution (20 µL, pH 7.4) was added to each well. Next, the plates were incubated for 4 hours. After careful extraction, the culture supernatant from each well was subjected to centrifugation. Next, following the addition of dimethyl sulfoxide (DMSO) (150 µL) to each well, the plates were then subjected to agitation for 10 minutes to ensure complete crystal dissolution. An ELISA (enzyme-linked immunosorbent assay) reader was utilized to measure absorbance at 490 nm for each well, and the obtained values were recorded. The growth curve of the cells was plotted with time and absorbance values on the X- and Y-axes, respectively.
Flow cytometry was performed to analyze cell cycle and apoptosis. To conduct the cell cycle analysis, cells (1×106/mL) were fixed with 70% ethanol for 1 h and then incubated for 30 minutes with a PI/RNase staining solution (CY001; Simu Biotech Co., Ltd., Tianjin, China). To determine cell apoptosis, cells (1×106/mL) were incubated with Annexin V-FITC and 7-AAD (A5001-03A; Simu Biotech Co., Ltd.) for 15 minutes. Subsequently, cell cycle and apoptosis were quantified by a flow cytometer (BD FACSVerse; Becton, Dickinson and Company, Franklin Hoo, NJ, USA).
The wound healing assay was conducted as follows. Treated glioma cells were seeded onto 6-well plates at 2×105 cells/mL density and grown till cell confluence was reached. The fusion monolayer was acquired. An incision was then created with one end of a sterile 200 µL pipette. After washing twice with phosphate buffer saline (PBS) to eliminate any cellular waste, the cells were incubated with a fresh solution of DMEM containing 3% FBS. The cells were subsequently cultured for 24 hours. The wound healing area was recorded using a microphotography technique.
In the Transwell invasion experiment, following 24-hour treatment, glioma cells were coated with a matrix (Millipore, Boston, MA, USA) at the density of 25 mg/cm2 in the upper chamber. After freezing the matrigel, 5.0×104 cells of each group were seeded onto each chamber containing a serum-free medium. Furthermore, 500 µL of serum-free medium was added to the lower chamber, which functioned as a chemical attractant. After incubation for 48 hours, noninvasive cells were gently removed from the upper chamber by using a cotton swab. The cells on the lower surface were fixed with 4% PFA instead of exposure to the medium in the bottom chamber. Following fixation for 15 minutes at RT, the samples were subjected to rinsing with PBS and subsequently subjected to crystal violet (0.2% solution) staining for 10 mintues. Ten image fields were quantified for each experimental condition.
At 24 hours before treatment, glioma cells were seeded onto 96-well plates and then subjected to washing with PBS. The Fura-4 calcium ion detection kit (Biyuntian Biological Reagent, Shanghai, China) was used for further cell treatment. After the incubation period, fluorescein-labeled enzyme was used for detection (Fluo-4AM shows green fluorescence) with a fluorometer at the dual excitation wavelengths of 490 nm and 525 nm.
Glioma cells treated under various conditions were cultured on glass covers for 24 hours. After thorough cleaning, treated cells were subjected to fixation with 4% PFA for 25 minutes. Subsequently, Triton X-100 (0.5%) was then used for cell permeabilization for 5 minutes. After incubation for 1 hour in a 5% BSA solution in PBS, the cells were then subsequently incubated with tubulin (mouse) antibodies (1 : 100 dilution) at 4°C overnight. Following washing for three times with PBS, the cells were incubated with Alexa 488-conjugated goat anti-rabbit secondary antibodies (1 : 10000) for 1 hour at 37°C. After incubation, the cells were washed with PBS and stained with DAPI, followed by DNA staining. An Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan; Center for Basic Medical Research, Tianjin Medical University) was used for observing the stained cells.
SPSS ver. 27.0 software (SPSS Inc., Chicago, IL, USA) was utilized for data analysis. analysis of variance, the least significant difference test, and Pearson’s correlation test were used for statistical analyses. The results are expressed as mean±standard error of mean. A p-value of <0.05 was considered statistically significant. The in vitro experiment was replicated thrice.
A neuropathologist histologically evaluated glioma and normal brain tissue specimens. p120 expression in gliomas was assessed by immunohistochemical staining and western blotting assay. Immunohistochemical analysis of the tissue sections revealed cytoplasmic localization of p120, with a positive correlation of its signal intensity with the malignancy grade of tumors (Fig. 1A). The p120 expression level in nontumor brain tissues was 14.50%±1.54% (normal brain tissue; n=10). In accordance with the WHO grading system, the p120 expression level was increased to 43.00%±3.81% in grade I tumors (n=6), further elevated to 59.17%±2.98% in grade II tumors (n=10), significantly increased to 88.67%±5.30% in grade III tumors (n=9), and reached the highest level at 99.60%±6.31% in grade IV tumors (n=17) (Fig. 1B and C). The quantitative PCR was used to evaluate the CTNND1 expression levels in normal brain tissues and glioma tissues of different grades. The relative transcript level in non-tumor brain tissues was 1.00±0.02 (NB, n=13). The relative transcript level was 2.12±0.07 in grade II tumors (n=23), 3.76±0.10 in grade III tumors (n=25), and 4.39±0.23 in grade IV tumors (n=23) (Fig. 1D). Thus, glioma brain tissues showed a remarkable elevation of p120 expression as compared to nontumor brain tissues (p=0.00).
In the three glioma cell lines, as observed in western blotting assay, siRNA-1 and siRNA-2 groups exhibited a remarkable reduction in p120 expression as compared to siRNA-BC and siRNA-NC groups (p<0.05) (Fig. 2). Compared to the expression level of β-actin, siRNA-1 and siRNA-2 groups showed a significant decline in p120 expression (18.89%±1.70% and 26.85%±2.50% in LN229; 18.45%±1.65% and 15.57%±1.38% in U251; and 12.81%±1.20% and 9.71%±0.89% in U87 cells, respectively) as compared to the siRNA-NC group (79.95%±4.86% in LN229, 84.36%±5.71% in U251, and 73.03%±4.01% in U87 cells) and the siRNA-BC group (80.44%±4.88% in LN229, 87.34%±5.74% in U251, and 80.08%±4.90% in U87 cells).
The MTT method, also referred to as MTT colorimetry, is used to assess cell viability and proliferation. The underlying principle involves the reduction of exogenous MTT by succinate dehydrogenase in the mitochondria of viable cells, resulting in the formation and deposition of water-insoluble blue-purple formazan crystals in the cells. Nonviable cells lack this functionality. DMSO can dissolve the formazan crystals present in the cells. Subsequently, an ELISA is performed to determine the light absorption value at 490 nm wavelength. This measurement indirectly reflects the quantity of living cells (Fig. 3) (p<0.05).
Flow cytometry is a common method to measure cell apoptosis. The method is based on the following principle : cell apoptosis increases the permeability of the cell membrane; however, this increase in cell permeability varies between normal cells and necrotic cells. Based on this feature, the examined cell suspension was stained with fluorescein, and the fluorescence intensity in the cell suspension was estimated by flow cytometry to differentiate normal cells from necrotic cells and apoptotic cells. The apoptotic ability of the glioma cells increased remarkably after siRNA treatment (Fig. 4) (p<0.05).
In vitro cell migration was analyzed by the wound healing experiment. Although this method does not exactly replicate in vivo cell migration, it partially simulates the migratory behavior of cells during wound healing. To examine siRNA-induced inhibition of cortical migration, we conducted experiments with human glioma cells assigned to different treatment groups. At 0 and 24 hours of wound creation, wound closure was assessed through micrographs. The wound healing areas in siRNA-1 and siRNA-2 groups (15.45%±1.53%, 16.44%± 1.49% in LN229; 28.78%±2.63% and 29.18%±2.79% in U251; 13.93%±1.13% and 15.71%±1.54% in U87 cells) were smaller than those in the siRNA-NC group (32.69%±2.84% in LN229, 53.87%±3.70% in U251, and 43.00%±3.21% in U87 cells) and the siRNA-BC group (31.89%±3.11% in LN229, 54.31%± 3.75% in U251, and 44.36%±3.16% in U87 cells). Thus, siRNA-mediated p120 expression inhibition could effectively decrease glioma cell ability to migrate (Fig. 5) (p<0.05).
The invasiveness of tumor cells was determined using Transwell chambers containing Matrigel. Because cell invasion and migration are crucial features associated with diffuse glioma growth, we conducted a Transwell invasion assay to elucidate how p120 siRNA inhibition affected tumor cell invasiveness. The number of cells that exhibited invasive ability in the siRNA-1 and siRNA-2 groups (27.33%±2.33% and 30.07%±2.40% in LN229; 47.33%±2.80% and 44.65%±2.66% in U251; 22.23%±1.76% and 22.68%±1.84% in U87) was less than those in the siRNA-NC group (83.89%±4.84% in LN229, 105.85%±5.71% in U251, and 87.67%±5.01% in U87 cells) and the siRNA-BC group (87.65%±4.93% in LN229, 102.32%±5.06% in U251, and 86.33%±4.97% in U87). These findings revealed that siRNA transfection significantly reduced the invasiveness of LN229, U251, and U87 glioma cells (Fig. 6) (p<0.05).
Calcium ions, as important regulatory factors in cells, play a crucial role in cellular biological activities. Calcium overload is a prominent manifestation in the process of cell apoptosis. Following p120 knockdown in glioma cells, both siRNA-treated groups exhibited a significant increase in the intracellular calcium ion concentration (Fig. 7) (p<0.05).
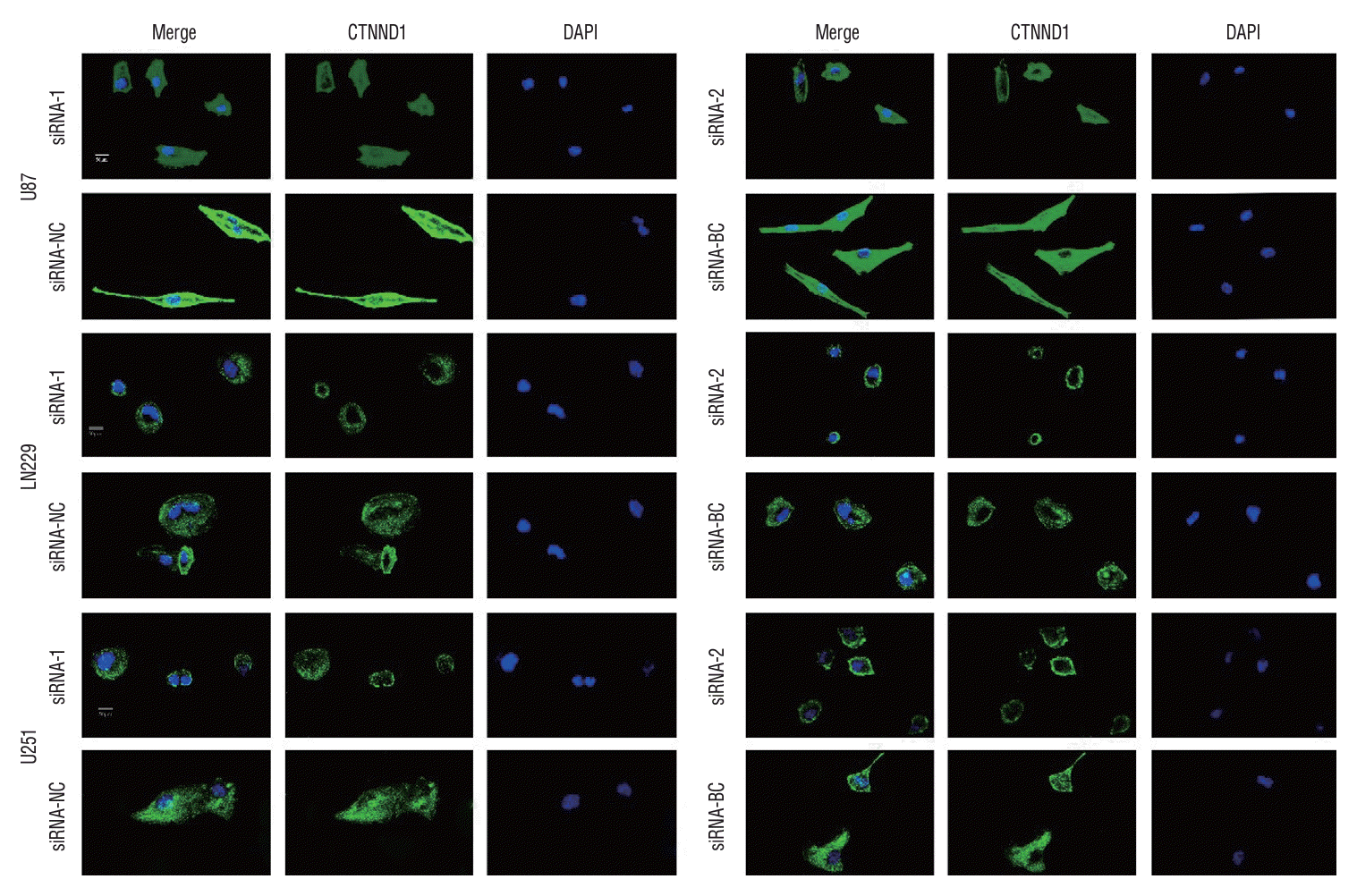
To examine whether p120-targeted siRNA suppresses microtubule assembly in glioma cells, we performed immunofluorescence anti-tubulin staining for microtubules and examined cell nuclei by DAPI staining. The findings indicate a notable decline in glioma cell capacity to form microtubules following p120 downregulation. This implies that p120 has a crucial involvement in facilitating microtubule formation in glioma cells (Fig. 8).
Glioma, a common intracranial malignant tumor, has high morbidity, mortality, and disability rates. Despite the availability of multiple treatment approaches, including surgery, radiotherapy, and chemotherapy, achieving a definitive cure is challenging as the malignancy tends to escalate upon recurrence [1,37]. Despite advancements in therapeutic modalities, such as the application of electromagnetic fields, the prognosis of glioma remains poor [22,24]. Various studies have shown that TMs, as an important part of the glioma TME, significantly promote the malignant effects of glioma [23,33,35].
As a protein closely related to cell movement and intercellular adhesion, p120 has received considerable attention in the past few years [27]. The involvement of p120 in the formation and regulation of TMs has been observed across various tumor cell types. Studies have demonstrated that tumor cells can effectively exchange small molecules, such as IP3, microRNAs, and adenosine-triphosphate, through specialized transport mechanisms, thereby ensuring intracellular energy supply and intercellular information transmission to facilitate the malignant biological behavior of tumor cells. The intertumor network established by TEMs can effectively regulate the distribution of toxic substances in a large number of glioma cells, thereby mitigating local toxicity levels, preventing calcium overload, and ultimately averting cellular death. It is therefore crucial to study p120 catenin involvement in the malignant characteristics of glioma and the potential underlying mechanism [10,18,34].
The formation of intercellular networks significantly enhances the progression of glioma and confers resistance to standard treatments, including surgery, chemotherapy, and radiotherapy [17,20,25]. Functional imaging of glioma cells following radiotherapy demonstrated a higher likelihood of cell death in individual glioma cells lacking network connections; this was attributed to intracellular calcium ion overload. In contrast, glioma cells exhibiting network connections showed reduced intracellular calcium ion concentrations. Consequently, the interconnections between tumors could potentially regulate intracellular calcium ion concentrations in glioma cells, thereby exerting a protective effect against cell apoptosis [19,28,29].
In this investigation, we initially utilized a range of experimental methodologies to confirm the connection between CTNND1 expression and glioma grade, with the results indicating a direct link between the level of expression and the severity of glioma. In the present study, we successfully acquired three types of common human glioma cells with low CTNND1 expression through RNA interference technology and utilized them as the subjects for subsequent in vitro experimental investigation. The well-established fact is that glioma cells, characterized by their uncontrolled proliferation and invasive growth, result in a significantly shortened median survival. Subsequently, we utilized a range of commonly employed and dependable experimental techniques to examine the changes in the malignant biological behavior of human glioma cells exhibiting low CTNND1 expression, encompassing cell proliferation, apoptosis, migration, and invasion. The results indicated that the down-regulation of CTNND1 led to a decreased ability of proliferation, apoptosis, migration, and invasion in glioma cells compared to the untreated group. The findings indicate that CTNND1 plays a pivotal role in the malignant biological behavior of human glioma cells. p120, identified as the upstream protein by GAP43, is believed to be implicated in the development of glioma intercellular microtubules (TMs) and thus linked to intracellular calcium ion exchange, although this remains unconfirmed. The study’s results revealed that CTNND1 is responsible for the reduction of calcium levels within progenitor glioma cells, resulting in an overload of cellular calcium and heightened susceptibility to apoptosis [2,26]. Finally, we have determined that CTNND1 negatively regulates tubulin levels and microtubule formation in descendant glioma cells, thus validating the association between CTNND1 and TMs formation in vitro. The overall experimental results demonstrated that p120 (CTNND1) exerts a positive influence on the malignant biological behavior of glioma, potentially by promoting the formation of microtubules in glioma cells, facilitating material exchange between these cells, and enhancing cell invasion. p120 (CTNND1) is a vital protein involved in the formation of intercellular connections within glioma cells, and it requires additional investigation, including experiments conducted in vivo. It has the potential to be a target for glioma therapy.
Notes
Informed consent
Informed consent was obtained from all individual participants included in this study.
References
1. Barthel L, Hadamitzky M, Dammann P, Schedlowski M, Sure U, Thakur BK, et al. Glioma: molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 41:53–75. 2022.

2. Becchetti A. Chapter two - interplay of Ca2+ and K+ signals in cell physiology and cancer. Curr Top Membr. 92:15–46. 2023.
3. Boiarska Z, Passarella D. Microtubule-targeting agents and neurodegeneration. Drug Discov Today. 26:604–615. 2021.
4. Di Cristofori A, Basso G, de Laurentis C, Mauri I, Sirtori MA, Ferrarese C, et al. Perspectives on (a)symmetry of arcuate fasciculus. A short review about anatomy, tractography and TMS for arcuate fasciculus reconstruction in planning surgery for gliomas in language areas. Front Neurol. 12:639822. 2021.
5. Duñach M, Del Valle-Pérez B, García de Herreros A. p120-catenin in canonical Wnt signaling. Crit Rev Biochem Mol Biol. 52:327–339. 2017.

6. Goodson HV, Jonasson EM. Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol. 10:a022608. 2018.

7. Haag D, Zipper P, Westrich V, Karra D, Pfleger K, Toedt G, et al. Nos2 inactivation promotes the development of medulloblastoma in Ptch1(+/-) mice by deregulation of Gap43-dependent granule cell precursor migration. PLoS Genet. 8:e1002572. 2012.

8. Hatanaka K, Simons M, Murakami M. Phosphorylation of VE-cadherin controls endothelial phenotypes via p120-catenin coupling and Rac1 activation. Am J Physiol Heart Circ Physiol. 300:H162–H172. 2011.

9. Hernández A, Domènech M, Muñoz-Mármol AM, Carrato C, Balana C. Glioblastoma: relationship between metabolism and immunosuppressive microenvironment. Cells. 10:3529. 2021.

10. Hong JY, Oh IH, McCrea PD. Phosphorylation and isoform use in p120-catenin during development and tumorigenesis. Biochim Biophys Acta. 1863:102–114. 2016.

11. Jiang G, Wang Y, Dai S, Liu Y, Stoecker M, Wang E, et al. P120-catenin isoforms 1 and 3 regulate proliferation and cell cycle of lung cancer cells via β-catenin and Kaiso respectively. PLoS One. 7:e30303. 2012.

12. Jiang Y, Liao L, Shrestha C, Ji S, Chen Y, Peng J, et al. Reduced expression of E-cadherin and p120-catenin and elevated expression of PLC-γ1 and PIKE are associated with aggressiveness of oral squamous cell carcinoma. Int J Clin Exp Pathol. 8:9042–9051. 2015.
13. Jung E, Osswald M, Ratliff M, Dogan H, Xie R, Weil S, et al. Tumor cell plasticity, heterogeneity, and resistance in crucial microenvironmental niches in glioma. Nat Commun. 12:1014. 2021.

14. Krigers A, Demetz M, Moser P, Kerschbaumer J, Brawanski KR, Fritsch H, et al. Impact of GAP-43, Cx43 and actin expression on the outcome and overall survival in diffuse and anaplastic gliomas. Sci Rep. 13:2024. 2023.

15. Latario CJ, Schoenfeld LW, Howarth CL, Pickrell LE, Begum F, Fischer DA, et al. Tumor microtubes connect pancreatic cancer cells in an Arp2/3 complex-dependent manner. Mol Biol Cell. 31:1259–1272. 2020.

16. Lee M, Ji H, Furuta Y, Park JI, McCrea PD. p120-catenin regulates REST and CoREST, and modulates mouse embryonic stem cell differentiation. J Cell Sci. 127(Pt 18):4037–4051. 2014.
17. Lou E. A ticket to ride: the implications of direct intercellular communication via tunneling nanotubes in peritoneal and other invasive malignancies. Front Oncol. 10:559548. 2020.

18. Maiden SL, Petrova YI, Gumbiner BM. Microtubules inhibit E-cadherin adhesive activity by maintaining phosphorylated p120-catenin in a colon carcinoma cell model. PLoS One. 11:e0148574. 2016.

19. Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 528:93–98. 2015.

20. Osswald M, Solecki G, Wick W, Winkler F. A malignant cellular network in gliomas: potential clinical implications. Neuro Oncol. 18:479–485. 2016.

21. Parato J, Bartolini F. The microtubule cytoskeleton at the synapse. Neurosci Lett. 753:135850. 2021.

22. Poff A, Koutnik AP, Egan KM, Sahebjam S, D’Agostino D, Kumar NB. Targeting the Warburg effect for cancer treatment: ketogenic diets for management of glioma. Semin Cancer Biol. 56:135–148. 2019.

23. Portela M, Mitchell T, Casas-Tintó S. Cell-to-cell communication mediates glioblastoma progression in Drosophila. Biol Open. 9:bio053405. 2020.
24. Przybylowski CJ, Hervey-Jumper SL, Sanai N. Surgical strategy for insular glioma. J Neurooncol. 51:491–497. 2021.

25. Roehlecke C, Schmidt MHH. Tunneling nanotubes and tumor microtubes in cancer. Cancers (Basel). 12:857. 2020.

26. Saberbaghi T, Wong R, Rutka JT, Wang GL, Feng ZP, Sun HS. Role of Cl-channels in primary brain tumour. Cell Calcium. 81:1–11. 2019.
27. Venhuizen JH, Jacobs FJC, Span PN, Zegers MM. P120 and E-cadherin: double-edged swords in tumor metastasis. Semin Cancer Biol. 60:107–120. 2020.

28. Venkataramani V, Schneider M, Giordano FA, Kuner T, Wick W, Herrlinger U, et al. Disconnecting multicellular networks in brain tumours. Nat Rev Cancer. 22:481–491. 2022.

29. Venkataramani V, Yang Y, Schubert MC, Reyhan E, Tetzlaff SK, Wißmann N, et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 185:2899–2917.e31. 2022.

30. Venkatesh VS, Lou E. Tunneling nanotubes: a bridge for heterogeneity in glioblastoma and a new therapeutic target? Cancer Rep (Hoboken). 2:e1185. 2019.

31. Wang X, Liang J, Sun H. The network of tumor microtubes: an improperly reactivated neural cell network with stemness feature for resistance and recurrence in gliomas. Front Oncol. 12:921975. 2022.

32. Watson DC, Bayik D, Storevik S, Moreino SS, Sprowls SA, Han J, et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat Cancer. 4:648–664. 2023.

33. Weil S, Osswald M, Solecki G, Grosch J, Jung E, Lemke D, et al. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol. 19:1316–1326. 2017.

34. Wu H, Yan X, Zhao L, Li X, Li X, Zhang Y, et al. p120-catenin promotes innate antiviral immunity through stabilizing TBK1-IRF3 complex. Mol Immunol. 157:8–17. 2023.

35. Xie R, Kessler T, Grosch J, Hai L, Venkataramani V, Huang L, et al. Tumor cell network integration in glioma represents a stemness feature. Neuro Oncol. 23:757–769. 2021.

36. Xie Z, Tang Y, Man MQ, Shrestha C, Bikle DD. p120-catenin is required for regulating epidermal proliferation, differentiation, and barrier function. J Cell Physiol. 234:427–432. 2018.

Fig. 1.
Expression of p120 in human glioma specimens. A : The cytoplasmic localization of CTNND1, as indicated by the presence of brown staining observed and photographed under a microscope, exhibited a positive association with the malignancy level of glioma. B : Western blot analysis was employed to assess the presence of the two proteins in healthy brain tissues and various stages of glioma tissues (p<0.05). C : The Pearson correlation test was conducted to analyze the association between CTNND1 expression and the malignancy level of glioma specimens. The findings revealed a positive correlation between CTNND1 expression and the degree of malignancy in glioma (r=0.906; ****p<0.0001). D : The results of quantitative PCR indicated a high expression of CTNND1 in gliomas, which was positively associated with the grade of glioma (F=830.6, °p<0.01). NB : normal brain tissue, WHO : World Health Organization, CTNND1 : catenin (cadherin-associated protein), delta 1.
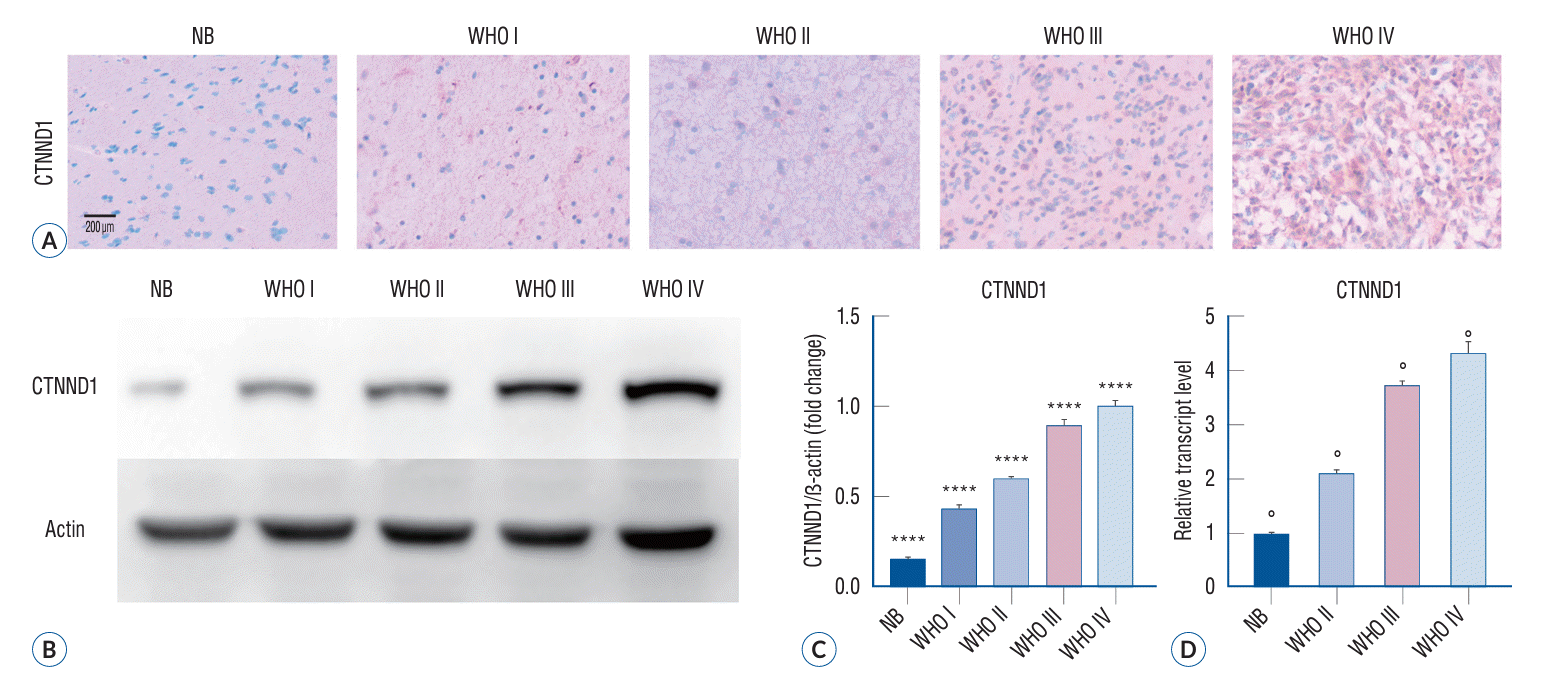
Fig. 2.
The expression of p120 in different treatment groups of glioma cells. With GAPDH as internal reference, the expression of p120 in siRNA-1 and siRNA-2 groups was significantly lower than that in siRNA-BC and siRNA-NC groups (A-D) (****p<0.0001). BC : blank control group, NC : negative control group, CTNND1 : catenin (cadherin-associated protein), delta 1, ns : no significance, GAPDH : glyceraldehyde-3-phosphate dehydrogenase.
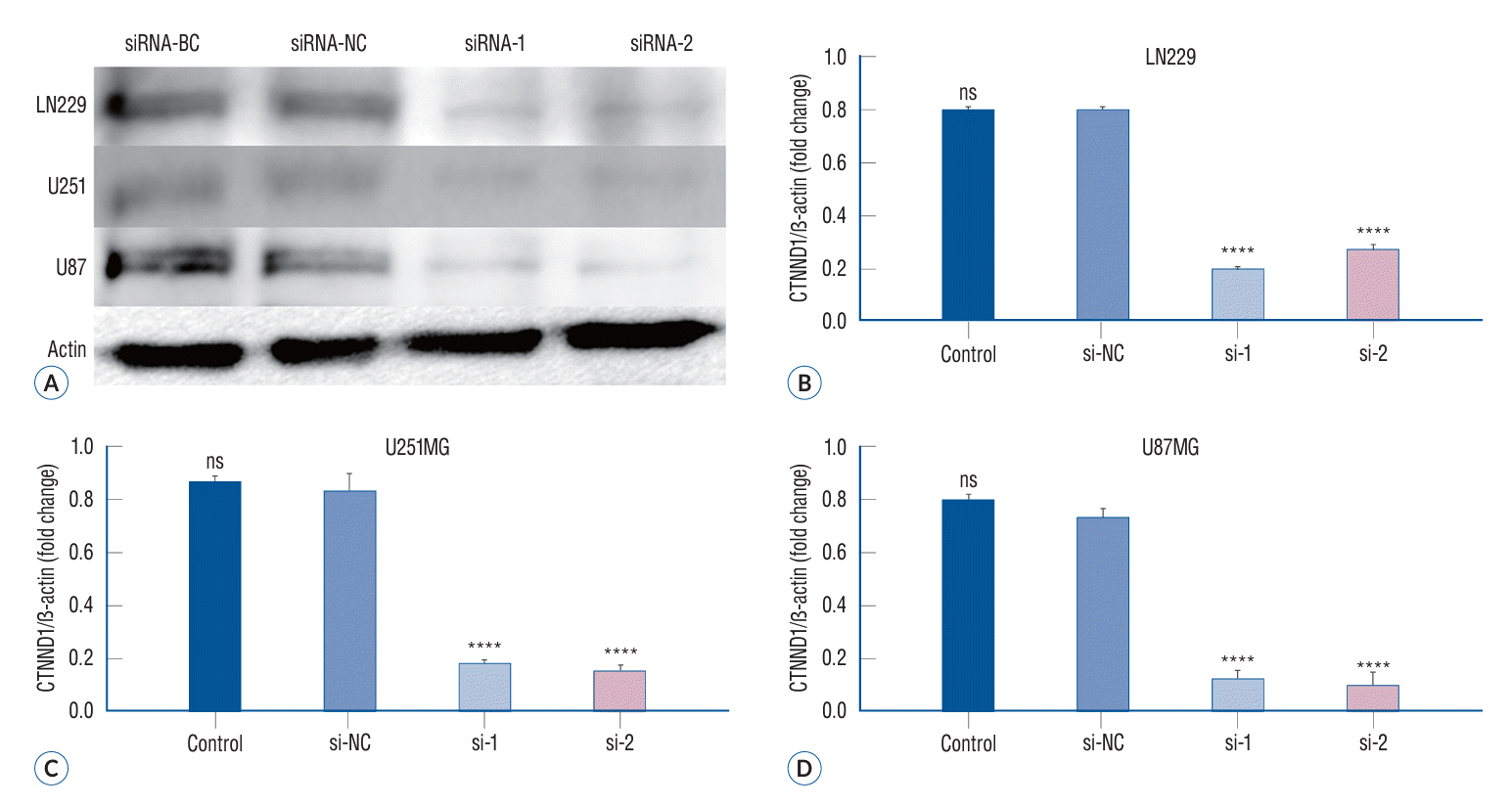
Fig. 3.
The proliferation of glioma cells in different treatment groups was evaluated by MTT assay. The MTT method, also referred to as MTT colorimetry, is a technique utilized for assessing cell viability and proliferation. The underlying detection principle involves the reduction of exogenous MTT by succinate dehydrogenase in the mitochondria of viable cells, resulting in the formation and deposition of water-insoluble blue-purple crystal Formazan within the cells. Conversely, non-viable cells lack this functionality. Dimethyl sulfoxide can dissolve the Formazan present in the cells, and subsequently, an enzyme-linked immunosorbent assay determines the light absorption value at either 540 or 720 nm wavelengths. The results showed that the proliferation ability of the two glioma cells decreased significantly after siRNA in the three malignant glioma cell lines (A-C). **p<0.01. ***p<0.001. ****p<0.001. shbc : siRNA blank controlshnc, shnc : siRNA normal control, sh1 : siRNA-1 group, sh2 : siRNA-2 group, ns : no significance, MTT : 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-diphenytetrazoliumromide.
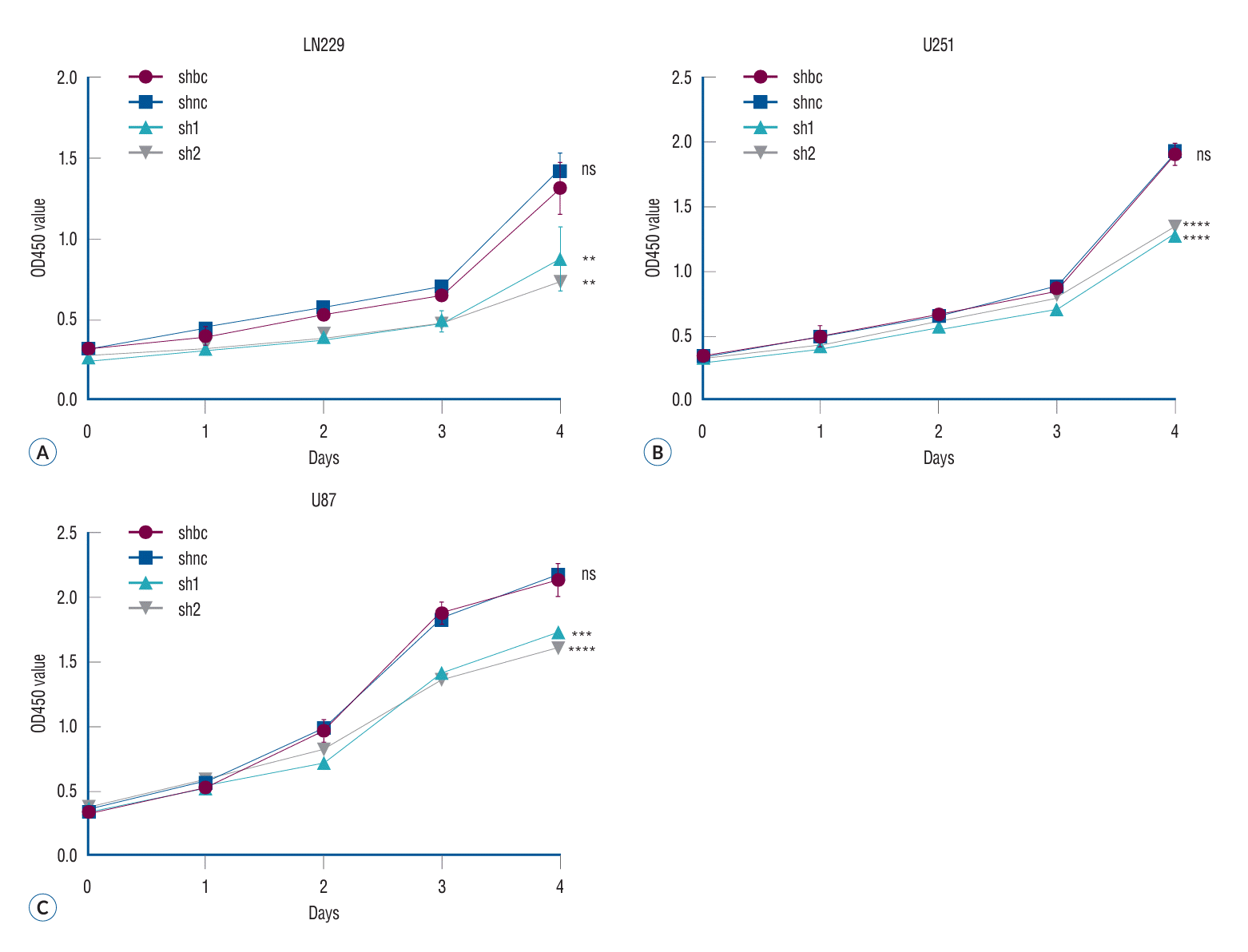
Fig. 4.
The apoptosis of glioma cells in different treatment groups was detected by flow cytometry. Flow cytometry is a common method to measure apoptosis. The principle is that when apoptosis occurs, the permeability of the cell membrane also increases, but the degree is between normal cells and necrotic cells. Using this feature, the examined cell suspension was stained with fluorescein, and the fluorescence intensity in the cell suspension was measured by flow cytometry to distinguish normal cells from necrotic cells and apoptotic cells. The results showed that the apoptosis ability of the two glioma cells increased significantly after siRNA treatment (A-C). *p<0.05. **p<0.01. ***p<0.001. ****p<0.001. BC : blank control group, NC : negative control group, 7-AADA-A : 7-aminoactinomycin D apoptosis, APC-A : allophycocyanin apoptosis, ns : no significance.
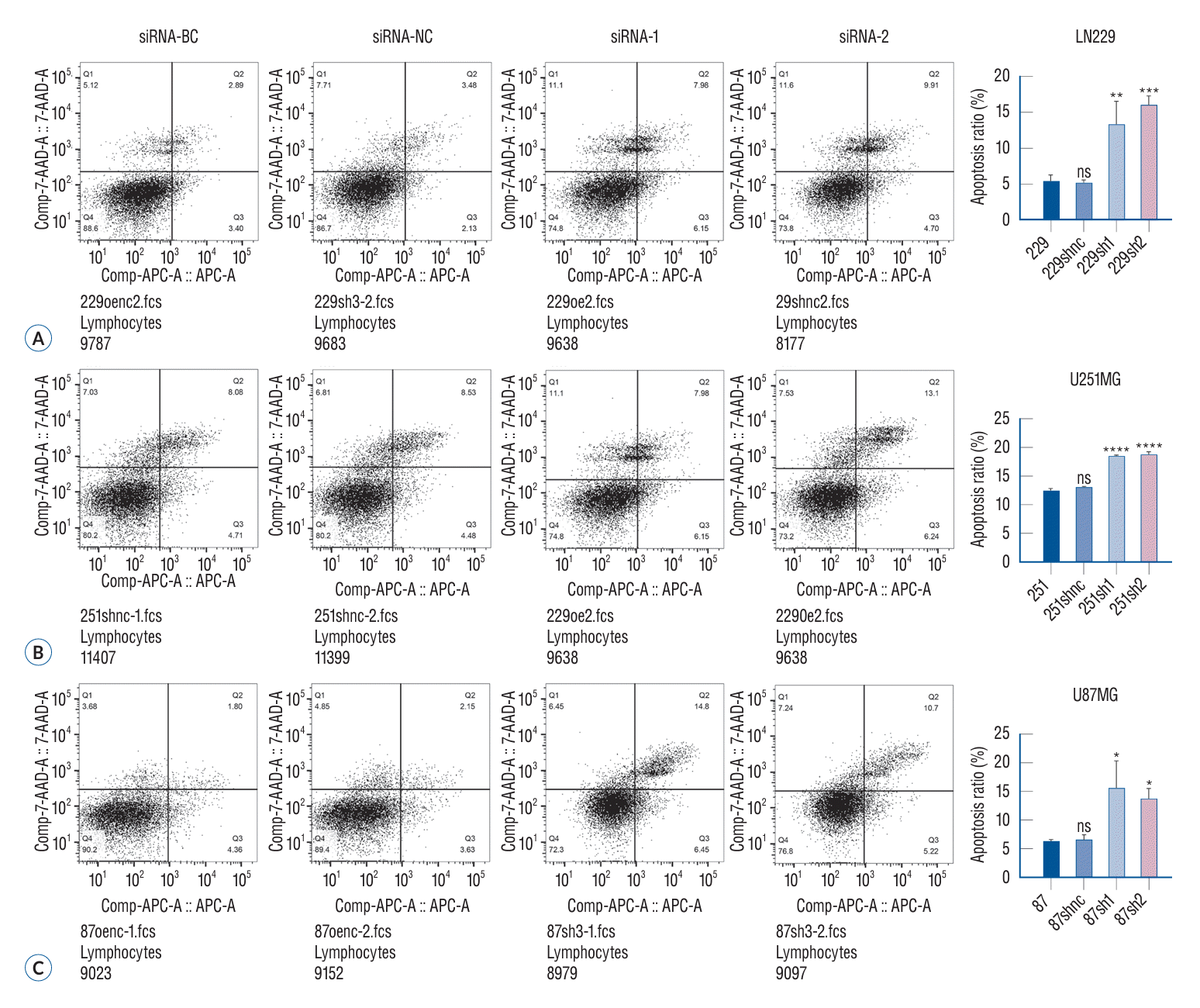
Fig. 5.
The migration ability of different treatment groups of glioma cells evaluated using a wound healing assay. After a 24-hour period of treatment involving the application of specific siRNA, a wound was created using the end of a sterile pipette with a volume capacity of 200 μL. Following this, the culture medium was substituted with serum-free medium and the cells were subsequently cultured for an additional 24 hours before being captured in photographs. A : In comparison to the siRNA-BC and siRNA-NC groups, the wound size was observed to be greater in the siRNA-1 and siRNA-2 groups 24 hours after the formation of wounds. B-D : In vitro experiments demonstrated a significant reduction in glioma cell migration upon treatment with P120-specific siRNA, as indicated by quantitative data (****p<0.0001). BC : blank control group, NC : negative control group, ns : no significance.
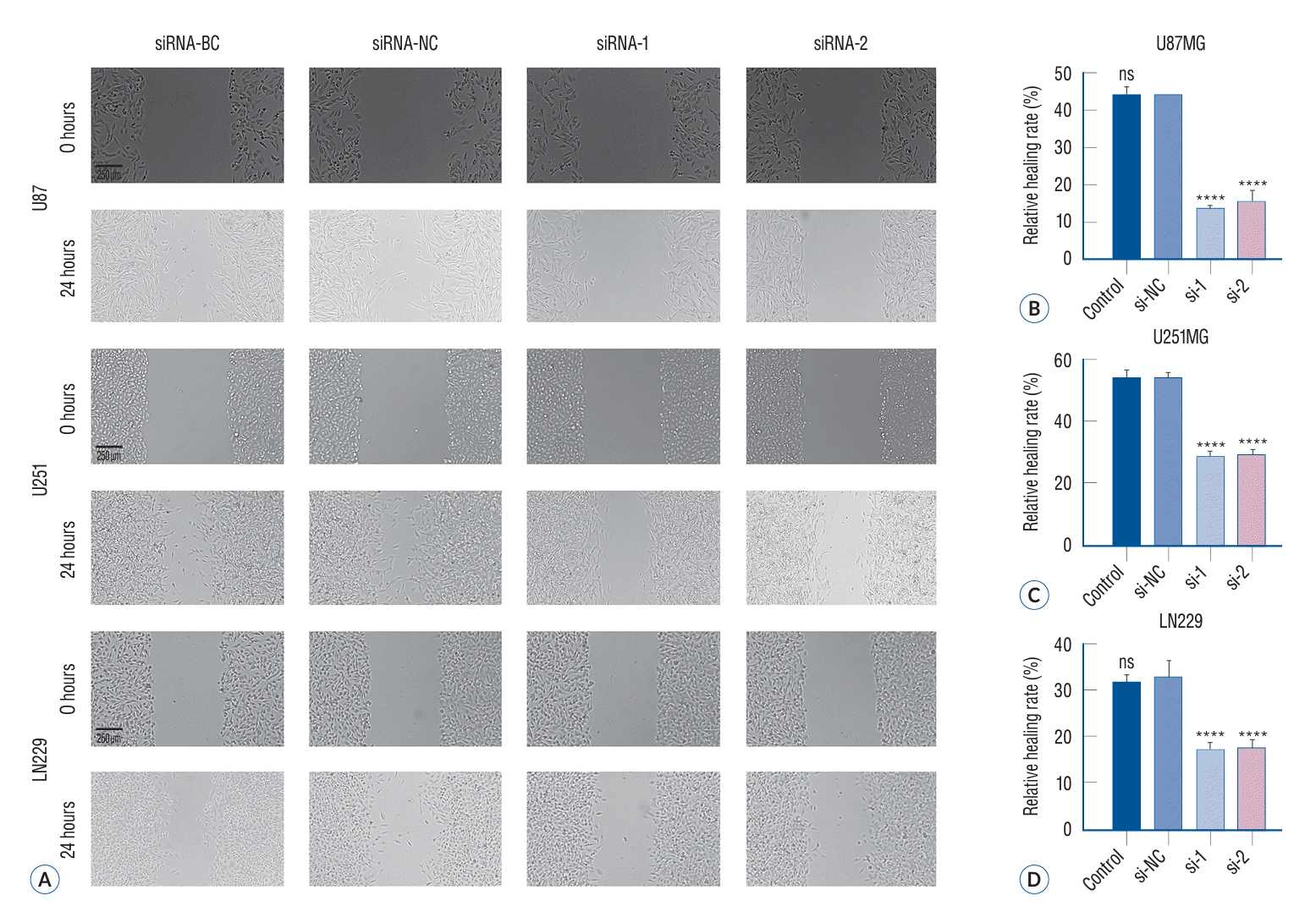
Fig. 6.
Knock down of cortactin inhibited glioma cell invasion in vitro. Following a 24-hour treatment period, we conducted a Transwell invasion assay and subsequently incubated the cells for an additional 48-hour. The stroma-migrating cells were then stained with crystal violet and photographed. Representative images of glioma cells in the Millipore Matrigel-coated invasion chambers (A). Relevant quantitative data demonstrated a significant reduction in the invasion of glioma cells in vitro upon specific siRNA treatment (B-D) (****p<0.0001). BC : blank control group, NC : negative control group, ns : no significance.
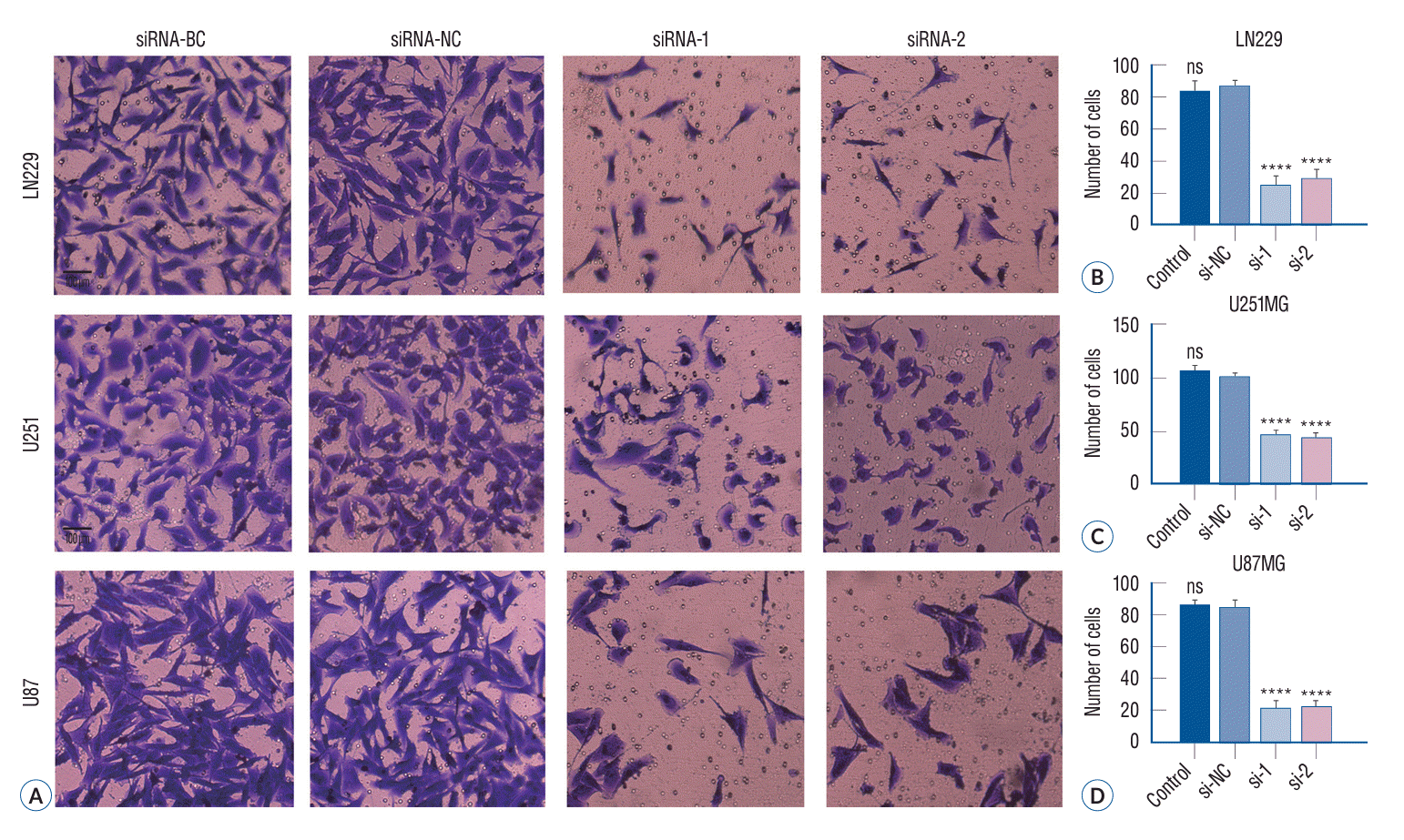
Fig. 7.
The concentration of calcium ion in glioma cells of different treatment groups was detected by enzyme-labeled method. Calcium channel is the channel through which cells transport calcium ions. Calcium overload caused by the increase of intracellular calcium ion concentration can cause the disorder of oxidative phosphorylation in mitochondria, the decrease of mitochondrial membrane potential, the decrease of adenosine-triphosphate content in tissues, and the activation of phospholipase and protease in cytoplasm, which can lead to and promote irreversible damage of cells. After the deletion of p120 expression in glioma cells, the intracellular calcium concentration in both groups was significantly increased (A-C). *p<0.05. ****p<0.0001.
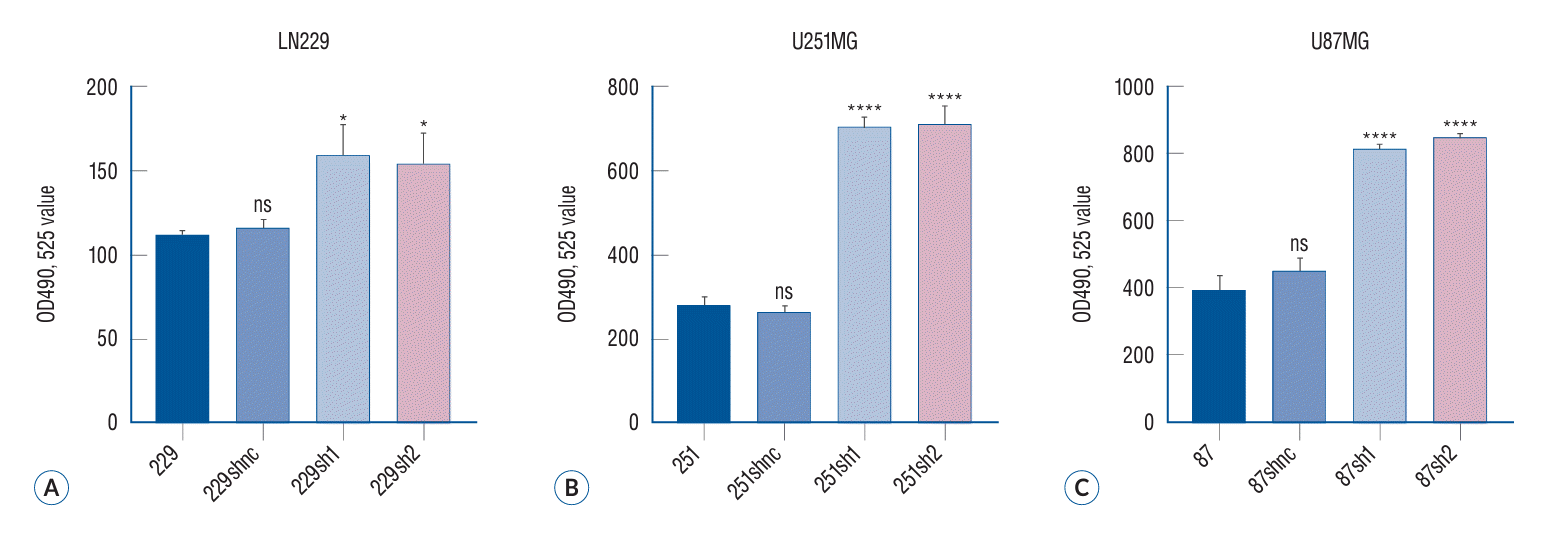
Fig. 8.
The microtubule formation of glioma cells in different treatment groups was detected by immunofluorescence. To assess the inhibitory impact of siRNA targeting p120 on microtubule assembly in glioma cells, we conducted immunofluorescence staining for tubulin and DAPI to examine the nuclei of different treatment groups. The findings revealed a significant reduction in the ability of glioma cells to form microtubules following downregulation of p120 expression. These outcomes imply that p120 plays a crucial role in facilitating microtubule formation within glioma cells. The maglification is ×400. CTNND1 : catenin (cadherin-associated protein), delta 1, DAPI : 4ʹ,6-diamidino-2-phenylindole, NC : negative control group, BC : blank control group.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download