Abstract
The evolving landscape of precision oncology underscores the pivotal shift from morphological diagnosis to treatment decisions driven by molecular profiling. Recent guidelines from the European Society for Medical Oncology recomend the use of next-generation sequencing (NGS) across a broader range of cancers, reflecting its superior efficiency and clinical value. NGS not only updates oncology testing by offering quicker, sample-friendly, and sensitive analysis but also reduces the need for multiple individual tests. Cytology samples, often obtained through less invasive methods, can yield high-quality genetic material suitable for molecular analysis. This article focuses on optimizing the use of cytology samples in NGS, and outlines their potential benefits in identifying actionable molecular alterations for targeted therapies across various solid tumors. It also addresses the need for validation studies and the strategies to incorporate or combine different types of samples into routine clinical practice. Integrating cytological and liquid biopsies into routine clinical practice, alongside conventional tissue biopsies, offers a comprehensive approach to tumor genotyping, early disease detection, and monitoring of therapeutic responses across various solid tumor types. For comprehensive biomarker characterization, all patient specimens, although limited, is always valuable.
The ongoing refinement of treatment guidelines in precision oncology represents a pivotal and unprecedented advancement in the field. The recent approval of molecularly targeted therapies has unveiled genomic signatures that are essential for guiding treatment decisions and enhancing the precision of outcome predictions. This rapid development points to a pivotal phase where multiplex genetic testing via next-generation sequencing (NGS) is increasingly recommended over single-gene testing (SGT). NGS is set to update oncology testing, offering greater efficiency and proven clinical value, thus reducing the reliance on numerous tests for individual gene alterations. The preference for NGS over SGT is strongly supported by emerging evidence.
The most recent European Society for Medical Oncology (ESMO) guidelines already reflect this shift, expanding the indications for NGS in patients with advanced tumors compared to the 2020 recommendations [1]. ESMO now recommended for the use of NGS in routine practice for advanced breast cancer, gastrointestinal stromal tumors, sarcoma, thyroid cancer, and cancer of unknown primary, in addition to the non–small cell lung cancer (NSCLC), cholangiocarcinoma, prostate cancer, and ovarian cancer, previously referred. Furthermore, ESMO recommends NGS for detecting tumor-agnostic alterations where matched therapies are available [2].
One of the major challenges associated with the implementation of NGS in the healthcare system is the cost and the difficulty in securing funding for testing the majority of patients at diagnosis and during tumor progression. However, the costs associated with NGS have significantly decreased in recent years. Moreover, data obtained from NGS not only predicts treatment response but also identifies resistance, potentially preventing the administration of unnecessary and expensive therapies. Supporting this direction, Stenzinger et al. [3] demonstrated that the cost per correctly identified patient was lower for NGS compared to sequential SGT in various cancer types, including advanced/metastatic non-squamous NSCLC, metastatic breast cancer, metastatic colorectal cancer, metastatic gastric cancer, and cholangiocarcinoma.
As the search for biomarkers predictive of responses to targeted therapies and immunotherapies continues to grow, it is crucial to consider biological samples beyond tissue specimens, such as cytological samples and liquid biopsies. Cytological examinations are increasingly used in routine oncology practice, as the demand for biomarker analysis, predictive of responses to targeted therapies and immunotherapies, continues to rise. Moreover, it is essential to explore the effective integration of these samples into molecular analysis procedures.
This article explores the key applications of cytological samples in next generation sequecing, focusing on their role in identifying actionable molecular alterations for targeted therapies. We also discuss next steps to integrate these samples into routine clinical practice, highlighting their potential as a valuable tool in precision cytopathology across major types of solid tumors, particularly in cases where cytological approach is already part of the clinical and diagnostic routine.
Most of the advances in predictive biomarkers have initially been based on studies and clinical trials that relied on histological specimens [4]. Accordingly, technical protocols and algorithms for evaluation or scoring had often been established for histological specimens, which raised the false impression that cytological specimens are not suitable for molecular testing by nature. This misconception among pathologists and clinicians was most prevalent in the early days of predictive marker testing and was reinforced by the popular but imprecise term “tissue is the issue.”
Cytology samples, in many cases, are the only available material for molecular testing [5], especially in situations where cancer diagnosis is made at late stage and surgery is not an option. These samples contain the same cells, the same RNA, DNA, and protein molecules as corresponding histology. Nonetheless, cytological specimens have no tissue context, usually lack a stromal component, and are processed differently, which may require different approaches and modified protocols for molecular analysis. Furthermore, cytology material involves a greater number of pre-analytical variables compared to standard formalin-fixed, paraffin-embedded (FFPE) tissue samples [6]. Nevertheless, any cytology sample, with adequate cellularity and preservation, may be used for molecular testing [7].
The pre-analytical factors that affect NGS analysis in cytology include type of preparation, type of fixative and stains, type of glass slides, specimen cellularity, tumor fraction, DNA yield, and input DNA [8]. Among these factors, studies frequently identify the low cellularity often encountered during sample preparation as one of the major obstacles to achieving molecular results with cytology specimens [9,10].
In molecular testing platforms like NGS, the required percentage of neoplastic cells depends on analytic sensitivity, with higher cellular content enhancing total DNA yield [6]. The percentage of neoplastic cells is more critical than the amount of DNA input. If only a small quantity of DNA from a few cancer cells is preferentially amplified, it might predominantly reflect non-neoplastic components, potentially resulting in false-negative outcomes [11]. The specific sample requirements vary depending on the target capture, gene panel, and sequencing platform used. For instance, Illumina NGS typically demands more cells and/or a higher DNA input compared to Ion Torrent NGS, making the latter potentially more suitable for cytopathology specimens [12]. While a standardized threshold for the required number of tumor cells is still not established, some authors recommend that tumor cellularity for NGS analysis should exceed two-fold the detection limit suggested by the molecular technique [13].
A broader standard aims for a neoplastic nucleus proportion of at least 10%, ideally 20% or more. However, in samples with high overall cellularity but a low tumor fraction, macrodissection is the tool used to delineate the tumor content. On the other hand, in samples with low cellularity but a high tumor fraction, adding more material through sample complementation is the recommended approach [5,14].
Although these cytological samples might have reduced cellularity, they present advantages in molecular testing by providing nucleic acids of higher quality, including greater purity, cellular yield, and tumor fraction. The quality of the genetic material obtained has proven to be a critical factor in detecting genomic alterations. Studies have shown that even with reduced cellularity and DNA concentrations, with DNA input below the manufacturer’s recommended limit of 10 ng, it is possible to detect target genomic alterations from cytological samples [15]. It is worth noting that nucleic acid extraction for NGS can also be achieved using archived stained cytological slides. In a study by Zannini et al. [16], DNA was successfully extracted in 68% of cases from cytological samples with moderate (100–500 representative cells) to high cellularity (> 500 representative cells), with an average concentration of 19.8 ng/μL.
Interlaboratory ring assay study included 14 institutions worldwide that performed multigene mutational assays at quantitative molecular reference specificities on quantitative cytological molecular reference specimens, showed highly reproducible results across all laboratories in detection of mutations down to 5% of allele frequency (AF), despite the difference in smears staining and sequencing practices. Most laboratories using NGS (78%) successfully detected the study’s mutations for 5% and 10% AF (10% and 20% neoplastic cells, respectively). The concordance decreased when analyzing mutations with AF less than 1%, highlighting the variation in threshold settings for variant calling across the institutions [17].
In thoracic oncology, where cytological sampling is increasingly common, it is essential to also account for its application in molecular analysis [18]. These samples are often obtained through less invasive procedures. A significant number of lung cancer cases are identified through fine needle aspiration (FNA) or exfoliative cytology specimens [7]. Therefore, developing effective strategies to prioritize and utilize these samples for diagnosis and biomarker research is crucial for effective lung cancer management.
For patients with advanced-stage NSCLC confirmed through histology or cytology, the current ESMO Clinical Practice Guidelines (2024) recommend molecular testing for predictive biomarkers including EGFR, KRAS (G12C), BRAF (V600), ERBB2 and MET (exon 14 skipping) mutations, MET amplifications, ALK, RET, ROS1, NTRK1/2/3, and NRG1 fusions [2]. Apart from predictive markers for approved treatments, several new lung cancer biomarkers are emerging, they concern mutations in PIK3CA, BRCA1/2, MAP2K1, and BRAF non-V600, and amplifications in human epidermal growth factor receptor 2 (HER2). Furthermore, some emerging biomarkers target genomic alterations in squamous NSCLC. Those are mostly fusions, mutations, and amplifications in FGFR [19]. The rapid expansion in the number of clinically significant biomarkers for advanced-stage NSCLC patients is anticipated to accelerate even further. To keep up with both current and emerging biomarkers, it is essential to integrate routine large-panel NGS [20]. Therefore, cytological samples must be suitable for this requirement.
A recent meta-analysis, which included 21 studies involving 1,175 patients, evaluated the potential of endobronchial ultrasound–guided transbronchial needle aspiration (EBUS-TBNA) as a method for routinely obtaining specimens for NGS analysis. EBUS-TBNA demonstrates a high yield (~80.9% to 91.4%) for NGS, reaffirming the suitability of utilizing EBUS-TBNA to retrieve specimens for NGS. Thus, the adequacy of EBUS-TBNA specimens is directly dependent on the DNA requirements used by individual laboratories. Nonetheless, the total amount of DNA extracted from EBUS-TBNA was 868.7 ng, which appears sufficient for most NGS panels, meeting the minimum input requirement of 10 ng, the recommended 50 ng, and 200 ng for larger panels [21]. Another noteworthy finding is that the success rate of EBUS-TBNA for NGS correlates with the number of passes performed. The recommendations propose collecting additional samples (≥3) beyond those required for diagnosing NSCLC for molecular analysis [7]. While there is no precise data on the exact number of passes required, increasing the number of passes may enhance the yield.
In our casuistics, the use of cytological samples from lung cancer for NGS is part of our routine practice. Over 1 year (unpublished data), 58% of the samples were cell blocks (CBs), and 38% were cytological smears, with an average tumor cellularity of 45%. Around 67% of the cytological samples showed at least one genomic alteration, and clinically actionable molecular targets were identified in 79% of the cases.
Endoscopic ultrasound–guided fine needle aspiration (EUS-FNA) is the primary technique for sampling and diagnosing lesions suspected to be pancreatic cancer, with ductal adenocarcinoma being the predominant pathological diagnosis [22]. The National Comprehensive Cancer Network (NCCN) guidelines recommend molecular analysis for patients with locally advanced or metastatic disease and suggest considering germline testing for all those diagnosed with pancreatic cancer. Molecular profiling and germline testing are important for both treatment and screening. Specifically, molecular profiling helps tailor chemotherapy regimens to individual patients, enhancing treatment effectiveness and avoiding unnecessary chemotherapy in patients who are unlikely to benefit from it [23].
A significant challenge in pancreatic ductal adenocarcinoma (PDAC) diagnosis lies in obtaining adequate material, particularly when it comes to the viability of extracted DNA and RNA from pancreatic tissues. Factors such as enzymatic degradation, hypocellularity, low tumor fractions, and preparation-related limitations contribute to these difficulties. Furthermore, up to 90% of PDAC cases are marked by a dense desmoplastic stroma, which adds another layer of complexity to the diagnostic process [24].
Key driver genes implicated in pancreatic cancer comprise AKT1, ALK, BRAF, CTNNB1, CDKN2A, DDR2, EGFR, ERBB2, ERBB4, FBX7, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NOTCH1, NRAS, PTEN, PIK3CA, SMAD4, STK11, and TP53. Of these, KRAS, TP53, and CDKN2A mutations are the most frequently observed and play a significant role in the initiation and progression of PDAC. The genomic alterations with level I/II targeted therapy according to ESMO Scale for Clinical Actionability of Molecular Targets include germline pathogenic or likely pathogenic variants in BRCA1/2, KRAS mutations (p.G12C), germline pathogenic or likely pathogenic variants in PALB2, and NRGS1 fusions [2]. Approximately 10% of patients with advanced PDAC have KRAS wild-type tumors. This subgroup is enriched with actionable therapeutic targets, such as NRG1 fusions and alternative mitogen-activated protein kinase pathway drivers like BRAFV600E mutations.
In the diagnostic field, studies have shown that NGS is effective in evaluating pancreatic cyst fluid collected via EUS-FNA. NGS assay is especially specific for detecting intraductal papillary mucinous neoplasms and mucinous cystic neoplasms through KRAS/GNAS mutations, while alterations in TP53/PIK3CA/PTEN are associated with advanced neoplasia. In one study, 626 samples were analyzed over 43 months, with NGS showing 89% sensitivity and 100% specificity for advanced neoplasia [25]. Another study tested 1,933 cysts, where combining NGS with cytopathology improved sensitivity to 93% and maintained 95% specificity, highlighting the clinical significance of genomic alterations in pancreatic cysts [26].
In our experience, we demonstrated that PDAC cellblock samples from EUS-FNA could be evaluated by NGS. Even at low concentrations, high-quality extracted DNA, provided valuable insights into the genetic alterations and possible therapeutic targets [27]. Similarly, Redegalli et al. [28] confirmed the feasibility of NGS with cytologic samples acquired via EUS-FNA from PDAC, showing that 88% of cases had closely matching mutational profiles between surgical specimens and identified actionable mutations.
Over the past decade, the use of molecular testing for assessing thyroid nodules has risen significantly. According to Huang et al., an analysis of 471,364 patients who underwent thyroid FNA between 2011 and 2021, the utilization increased from 0.01% to 10.1%, in 2021, with an immediate and deeper increase in molecular testing adoption after 2015 [29]. As mentioned, current ESMO guidelines expand the indications and recommend carrying out tumor NGS in patients with advanced thyroid cancer. The list of genomic alterations includes RET (mutations and fusions) and BRAF mutations (p. V600E) [2].
The molecular testing in FNA of thyroid nodules can also optimize patient management by reducing unnecessary surgery on benign nodules, clarifying diagnostic indeterminate in thyroid cytology, and addressing dilemmas between non-malignant findings and the clinical malignant nodules [30]. The BRAF V600E mutation is known for its high specificity—over 99%— for diagnosing thyroid cancer, particularly papillary thyroid carcinoma, therefore, the presence of the BRAF V600E mutation is important when cytomorphological results are inconclusive [31]. Furthermore, screening for tumor mutations should be conducted in patients with anaplastic thyroid cancer, as targeted therapy with BRAF inhibitors has demonstrated promising results in this rare and highly aggressive neoplasm for those with the BRAF V600E mutation [32].
In the context of neoplastic metastatic progression, cytological samples—whether obtained through lymph node metastasis FNA or aspiration of cavity fluids—represent a crucial substrate in clinical practice. These methods play a in sampling and monitoring metastatic evolution. The combination of these procedures with ancillary techniques, such as NGS, has proven to be reliable and is being explored in the analysis of metastatic effusions, particularly in cases of metastatic breast and ovarian cancers.
The ESMO guidelines also extend the recommendations to include tumor NGS for patients with advanced breast cancer. The genomic alterations ESCAT I/II include ERBB2 amplifications, PIK3CA mutations, germline/somatic BRCA 1/2, PTEN mutations/deletions, AKT mutations, PALB2 germline/pathogenic variants. It is advised to perform tumor NGS as standard care for hormone receptor–positive/HER2-negative advanced breast cancer. Testing should be conducted after endocrine therapy resistance to maximize the chances of identifying ESR1 mutations. However, these recommendations apply to the analysis of tumor (or plasma) samples [2].
Studies have compared the sensitivity of FNA to tumor sampling for detecting somatic mutations in primary and metastatic breast cancer. They observed no significant difference in total DNA yield between the two approaches. In fact, FNA offered better cellularity, a higher tumor fraction, and enhanced sequencing metrics compared to tumor samples, while also being minimally invasive [24,33].
Gornjec et al. [34] demonstrated that cytological samples are equivalent to tumor tissue in detecting BRCA1/2 mutations in high-grade serous tubo-ovarian carcinoma using NGS. In their study, BRCA1 and BRCA2 mutation analysis was conducted on cytological samples (ascites, pleural effusion, and enlarged lymph nodes) from 44 women with primary or recurrent high-grade ovarian cancer, showing 100% concordance with mutation results from histological samples [34]. Another study on ascitic fluid cytology in epithelial ovarian cancer found that cytological samples identified all oncogenic mutations present in surgically resected specimens, along with additional mutations not detected in tissue samples [35].
Perfoming multigene NGS in patients with advanced cancers for the assessment of tumor-agnostic targeted therapies using cytological samples, particularly in the setting of metastatic progression, is feasible and may provide substantial benefits to these patients. Current guidelines emphasize the evaluation of the following tumor-agnostic biomarkers: NTRK1, 2, 3 fusions, RET and FGFR1/2/3 fusions/mutations, BRAF V600E mutations, microsatellite instability–high, and high tumor mutation burden (TMB) [2,36,37].
A limitation in applying NGS assays, regardless of sample type, relates to the turnaround time (TAT), which is typically longer compared to SGT. The TAT for NGS can vary due to several factors, including the complexity of the panel, sample quality, and the efficiency of the laboratory workflow. However, NGS provides a more comprehensive genomic profile, which can compensate for the longer TAT by delivering critical insights for patient management within a single test.
In our clinical practice, particularly through discussions in molecular tumor boards, we have observed that while the extended TAT may pose a consideration, its impact is often offset by the richness of actionable data provided. This is especially relevant in cases where therapeutic decisions depend on a broader molecular understanding. Wolff et al. [38] also reported that targeted DNA- and RNA-based NGS approaches yield higher true-positive testing rates compared to sequential testing with multiple SGT. This more comprehensive strategy leads to more appropriate treatment decisions and improved patient survival outcomes.
Moreover, the use of cytological samples in NGS assays can significantly reduce TAT, primarily because cytological specimens streamline the workflow by eliminating steps required for FFPE tissue. Jager et al. [39] revealed that an innovative cytology workflow can shorten NGS TAT by as much as 30% compared to FFPE samples.
The application of comprehensive genomic analysis and deep targeted sequencing for assessing tumor TMB and whole genome amplification (WGA) faces several limitations, including constraints on routine clinical use and difficulties in obtaining sufficient tissue material. Studies suggest that TMB evaluation using circulating DNA and cytological samples at diagnosis may help predict druggable responses. In a pilot study, TMB assessment in CBs was shown to be technically feasible, with results closely matching those from histological specimens in terms of total reads, mapped reads, read lengths, and on-target reads (97.49% vs. 98.45%) [40]. Additionally, cytological smears displayed consistent mutation counts and TMB values at 5% and 10% variant AF thresholds compared to FFPE samples [41]. Using NGS of WGA, Amemiya et al. [42] demonstrated that drug-matched variants could be accurately identified from limited cytological specimens (10–20 tumor cells), yielding results similar to those obtained from FFPE samples.
In the realm of precision medicine, recent advancements are concentrating on maximizing the utility of cytology samples, aiming not just at diagnosis but also at increasing the availability of samples for molecular analyses. Consequently, a range of strategies and techniques are being developed to incorporate or combine different types of samples, with the objective of enhancing diagnostic precision and therapeutic decisions. When tissue biopsies and cytological specimens are both available, the decision to process these samples concurrently or sequentially depends on the clinical scenario and diagnostic objectives. Tissue biopsies are typically prioritized, as they generally offer higher cellularity. Nonetheless, cytological specimens can serve as strong alternatives when tissue is limited (complementing the tumor content) or unavailable. Liquid biopsies are especially used to monitor treatment response and disease progression. This strategy allows for cost-efficiency while maximizing the molecular information obtained from each sample type.
Reflex testing involves requesting biomarker assessment immediately following a morphological diagnosis, without needing a specific request from clinicians. Recent expert consensus recommends comprehensive reflex biomarker testing for all patients with a suitable diagnosis of NSCLC, regardless of disease stage [43]. This approach reduces the time between diagnosis and the initiation of targeted treatment [44]. To address this, implementing reflex molecular genotyping on cytology-based can provide a viable alternative. Study indicates that this method improves the success rate of DNA molecular testing and suggest that using liquid cytology from FNA or bronchoscopy rinses, stored in separate vials from FFPE samples, can help address the demand for tumor tissue [45].
Cytology material can provide a liquid biopsy testing option. In fact, liquid biopsy extends beyond plasma. Multiple studies have revealed that non-plasm body fluid, such as urine, pleural fluid, cerebrospinal fluid, and supernatants from cytology specimens, can provide circulating free DNA (cfDNA). These fluids, whether containing free-floating cfDNA or DNA encapsulated in exosomes, offer reliable genomic information with less invasiveness than traditional tissue biopsies [46].
Fluids collected near tumors or metastases often have higher concentrations of tumor DNA and a greater mutant allelic fraction due to active secretion and cell death, compared to blood samples. Analyzing multiple non-blood fluids alongside plasma-based methods can enhance sensitivity and offer a more detailed view of spatial tumor heterogeneity [47].
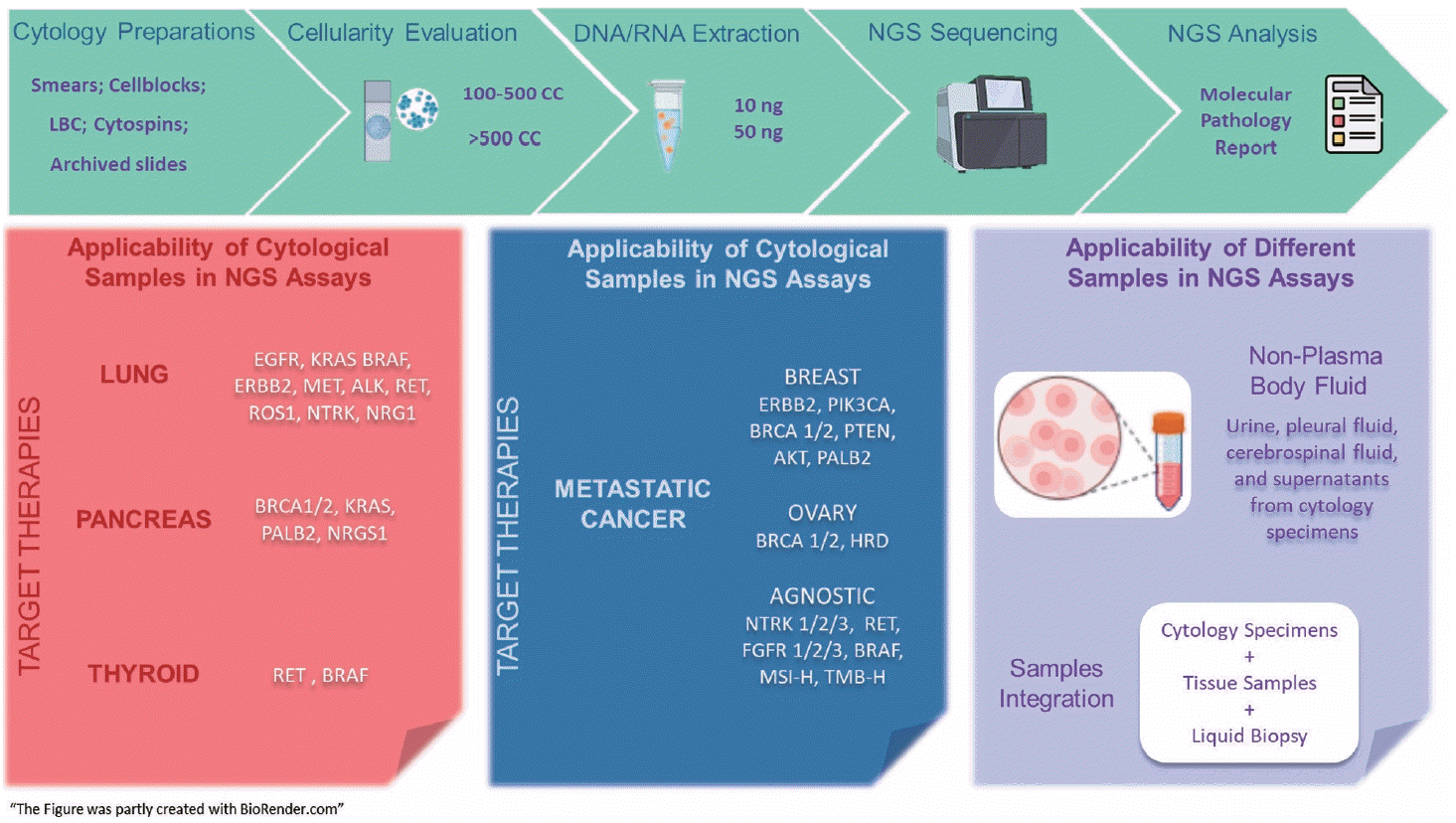
Replacing the primacy of morphological diagnosis by therapeutic decision, several efforts should be made to validate the analytical performance of the wide array of currently available molecular technologies, including multiplexed genetic testing. Multiplexed genetic testing with NGS is strongly recommended over SGT is quicker, sample-friendly and, sensitive. In clinical practice, all patient specimens are limited and valuable. The NGS assay supports the inclusion of conventional and innovative cytology sample preparations, supernatants, and body fluids (Fig. 1). Therefore, validation studies are essential, both intra- and inter-laboratory. Moreover, the integration of tissue biopsies, cytological specimens, and liquid biopsies is the way for the comprehensive characterization of current and emerging biomarkers. This strategy complements traditional techniques, enhancing tumor genotyping, early detection, monitoring of disease progression, and evaluating therapeutic response across main types of solid tumors.
Notes
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
References
1. Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020; 31:1491–505.

2. Mosele MF, Westphalen CB, Stenzinger A, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2021; 35:588–606.

3. Stenzinger A, Cuffel B, Paracha N, et al. Supporting biomarker-driven therapies in oncology: a genomic testing cost calculator. Oncologist. 2023; 28:e242–53.

4. Danesi R, Fogli S, Indraccolo S, et al. Druggable targets meet oncogenic drivers: opportunities and limitations of target-based classification of tumors and the role of Molecular Tumor Boards. ESMO Open. 2021; 6:100040.

5. Pisapia P, Pepe F, Sgariglia R, et al. Next generation sequencing in cytology. Cytopathology. 2021; 32:588–95.

6. Turner SA, Abou Shaar R, Yang Z. The basics of commonly used molecular techniques for diagnosis, and application of molecular testing in cytology. Diagn Cytopathol. 2023; 51:83–94.

7. Roy-Chowdhuri S, Dacic S, Ghofrani M, et al. Collection and handling of thoracic small biopsy and cytology specimens for ancillary studies: guideline from the College of American Pathologists in collaboration with the American College of Chest Physicians, Association for Molecular Pathology, American Society of Cytopathology, American Thoracic Society, Pulmonary Pathology Society, Papanicolaou Society of Cytopathology, Society of Interventional Radiology, and Society of Thoracic Radiology. Arch Pathol Lab Med. 2020; 144:933–58.

8. Morii E, Hatanaka Y, Motoi N, et al. Guidelines for handling of cytological specimens in cancer genomic medicine. Pathobiology. 2023; 90:289–311.

9. Sura GH, Tran K, Fu C, et al. Molecular testing opportunities on cytology effusion specimens: the pre-analytic effects of various body fluid cytology preparation methods on RNA extraction quality and targeted sequencing. J Am Soc Cytopathol. 2023; 12:10–9.

10. Hwang DH, Garcia EP, Ducar MD, Cibas ES, Sholl LM. Next-generation sequencing of cytologic preparations: an analysis of quality metrics. Cancer Cytopathol. 2017; 125:786–94.

11. Kim H, Chung JH. Biomarker testing of cytology specimens in personalized medicine for lung cancer patients. J Pathol Transl Med. 2022; 56:326–33.

12. Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing: the MD Anderson experience. Arch Pathol Lab Med. 2016; 140:1191–9.
13. Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.
14. Souza da Silva R, Pinto R, Cirnes L, Schmitt F. Tissue management in precision medicine: what the pathologist needs to know in the molecular era. Front Mol Biosci. 2022; 9:983102.

15. Roy-Chowdhuri S, Goswami RS, Chen H, et al. Factors affecting the success of next-generation sequencing in cytology specimens. Cancer Cytopathol. 2015; 123:659–68.

16. Zannini G, Tedesco I, Cozzolino I, et al. A critical issue in lung cancer cytology and small biopsies: DNA and RNA extraction from archival stained slides for biomarker detection through real time PCR and NGS: the experience in pathological anatomy unit. Diagnostics (Basel). 2023; 13:1637.

17. Malapelle U, Mayo-de-Las-Casas C, Molina-Vila MA, et al. Consistency and reproducibility of next-generation sequencing and other multigene mutational assays: a worldwide ring trial study on quantitative cytological molecular reference specimens. Cancer Cytopathol. 2017; 125:615–26.

18. Hofman P. What is new in biomarker testing at diagnosis of advanced non-squamous non-small cell lung carcinoma? Implications for cytology and liquid biopsy. J Mol Pathol. 2021; 2:147–72.

19. Hofman P, Berezowska S, Kazdal D, et al. Current challenges and practical aspects of molecular pathology for non-small cell lung cancers. Virchows Arch. 2024; 484:233–46.

20. de Jager VD, Timens W, Bayle A, et al. Future perspective for the application of predictive biomarker testing in advanced stage non-small cell lung cancer. Lancet Reg Health Eur. 2024; 38:100839.

21. Roh MH. The utilization of cytologic and small biopsy samples for ancillary molecular testing. Mod Pathol. 2019; 32:77–85.

22. Tempero MA, Malafa MP, Al-Hawary M, et al. Pancreatic adenocarcinoma, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021; 19:439–57.
23. Tempero MA, Malafa MP, Al-Hawary M, et al. Pancreatic adenocarcinoma, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021; 19:439–57.
24. Roy-Chowdhuri S, Chen H, Singh RR, et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for next-generation sequencing in solid organ malignancies. Mod Pathol. 2017; 30:499–508.

25. Singhi AD, McGrath K, Brand RE, et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut. 2018; 67:2131–41.

26. Paniccia A, Polanco PM, Boone BA, et al. Prospective, multi-institutional, real-time next-generation sequencing of pancreatic cyst fluid reveals diverse genomic alterations that improve the clinical management of pancreatic cysts. Gastroenterology. 2023; 164:117–33.

27. Souza da Silva R, Pina MJ, Cirnes L, Gouveia L, Albergaria A, Schmitt F. Comprehensive genomic studies on the cell blocks of pancreatic cancer. Diagnostics (Basel). 2024; 14:906.

28. Redegalli M, Schiavo Lena M, Cangi MG, et al. Proposal for a new pathologic prognostic index after neoadjuvant chemotherapy in pancreatic ductal adenocarcinoma (PINC). Ann Surg Oncol. 2022; 29:3492–502.

29. Huang Y, Chan SJ, Wright JD, et al. Does the adoption of molecular testing cause decreased thyroidectomy rates in a national cohort? A quasiexperimental study of high- versus low-adoption states. Thyroid. 2024; 34:388–98.

30. McMurtry V, Canberk S, Deftereos G. Molecular testing in fine-needle aspiration of thyroid nodules. Diagn Cytopathol. 2023; 51:36–50.
31. Schmitt F, da Silva RS. False positive in thyroid FNA: causes and how to avoid them. In: Kakudo K, Liu Z, Jung CK, Hirokawa M, Bychkov A, Lai CR, eds. Thyroid FNA cytology. Singapore: Springer Nature Singapore, 2023; 233-44.
32. Subbiah V, Kreitman RJ, Wainberg ZA, et al. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J Clin Oncol. 2018; 36:7–13.

33. Dupain C, Masliah-Planchon J, Gu C, et al. Fine-needle aspiration as an alternative to core needle biopsy for tumour molecular profiling in precision oncology: prospective comparative study of next-generation sequencing in cancer patients included in the SHIVA02 trial. Mol Oncol. 2021; 15:104–15.

34. Gornjec A, Novakovic S, Stegel V, et al. Cytology material is equivalent to tumor tissue in determining mutations of BRCA 1/2 genes in patients with tubo-ovarian high grade serous carcinoma. BMC Cancer. 2019; 19:296.

35. Nozaki T, Sakamoto I, Kagami K, et al. Molecular analysis of ascitic fluid cytology reflects genetic changes of malignancies of the ovary equivalent to surgically resected specimens. Cancer Cytopathol. 2022; 130:640–9.

36. Malapelle U, Pepe F, Pisapia P, et al. Reference standards for gene fusion molecular assays on cytological samples: an international validation study. J Clin Pathol. 2023; 76:47–52.

37. Schmitt F, Di Lorito A, Vielh P. Molecular testing on cytology for gene fusion detection. Front Med (Lausanne). 2021; 8:643113.

38. Wolff HB, Steeghs EMP, Mfumbilwa ZA, et al. Cost-effectiveness of parallel versus sequential testing of genetic aberrations for stage IV non-small-cell lung cancer in the Netherlands. JCO Precis Oncol. 2022; 6:e2200201.

39. Jager L, Jennings LJ, Dittmann D, Blanco J, Choy B, Nayar R. Supernatant fluid from endobronchial ultrasound-guided transbronchial needle aspiration for rapid next-generation sequencing. J Am Soc Cytopathol. 2024; 13:340–5.

40. Pepe F, Pisapia P, Gristina V, et al. Tumor mutational burden on cytological samples: a pilot study. Cancer Cytopathol. 2021; 129:460–7.

41. Alborelli I, Bratic Hench I, Chijioke O, et al. Robust assessment of tumor mutational burden in cytological specimens from lung cancer patients. Lung Cancer. 2020; 149:84–9.

42. Amemiya K, Hirotsu Y, Mochizuki H, et al. Deep targeted sequencing of cytological tumor cells using whole genome amplification. Cancer Cytopathol. 2023; 131:58–68.

43. Gosney JR, Paz-Ares L, Janne P, et al. Pathologist-initiated reflex testing for biomarkers in non-small-cell lung cancer: expert consensus on the rationale and considerations for implementation. ESMO Open. 2023; 8:101587.

44. Pisapia P, Pepe F, Iaccarino A, et al. Next generation sequencing in cytopathology: focus on non-small cell lung cancer. Front Med (Lausanne). 2021; 8:633923.

45. Marmarelis ME, Scholes DG, McGrath CM, et al. Brief report: impact of reflex testing on tissue-based molecular genotyping in patients with advanced non-squamous non-small cell lung cancer. Clin Lung Cancer. 2024; 25:262–5.

46. Souza da Silva R, Schmitt F. Minimally invasive, maximally effective: the power of precision cytoanalysis on effusion samples: a comprehensive exploration from traditional methods to innovative approaches. Surg Pathol Clin. 2024; 17:453–81.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download