Abstract
Objectives
This study evaluated the efficacy of integrating a retrieval-augmented generation (RAG) model and a large language model (LLM) to improve the accuracy of drug name mapping across international vocabularies.
Methods
Drug ingredient names were translated into English using the Japanese Accepted Names for Pharmaceuticals. Drug concepts were extracted from the standard vocabulary of OHDSI, and the accuracy of mappings between translated terms and RxNorm was assessed by vector similarity, using the BioBERT-generated embedded vectors as the baseline. Subsequently, we developed LLMs with RAG that distinguished the final candidates from the baseline. We assessed the efficacy of the LLM with RAG in candidate selection by comparing it with conventional methods based on vector similarity.
Results
The evaluation metrics demonstrated the superior performance of the combined LLM + RAG over traditional vector similarity methods. Notably, the hit rates of the Mixtral 8x7b and GPT-3.5 models exceeded 90%, significantly outperforming the baseline rate of 64% across stratified groups of PO drugs, injections, and all interventions. Furthermore, the r-precision metric, which measures the alignment between model judgment and human evaluation, revealed a notable improvement in LLM performance, ranging from 41% to 50% compared to the baseline of 23%.
Observational Health Data Sciences and Informatics (OHDSI) [1], the successor to the Observational Medical Outcomes Partnership (OMOP) project [2], has been working with cohorts comprising hundreds of millions of patients worldwide, to provide insights into real-world evidence. This global cross-sectional analysis is facilitated by converting medical information from various countries into a common data model structure, initially developed by OMOP, and by harmonizing medical terms using the OHDSI Standard Vocabulary (OSV) [3]; The goal is to standardize database queries. Before a country can participate in an OHDSI research project, its medical information structure and terminology must be aligned with the OSV [4,5]. Mapping concepts from a controlled vocabulary that has not been adopted as a source for OSV standard concepts requires considerable effort [6]. Additionally, drug-related terms require more consideration than those in other fields due to their complexity. The OSV incorporates international pharmaceutical terms through a proprietary system known as the RxNorm Extension, which builds on RxNorm [7], developed by the United States National Library of Medicine [3]. Thus, non-English-speaking countries need to translate drug names and pharmaceutical ingredients into English and align them with the corresponding concepts in RxNorm or the RxNorm Extension within the OSV. This alignment must take into account variations in dosage forms, quantities, and drug ingredients. To assist with this mapping, OHDSI offers the Usagi software tool, an official OSV mapper that aligns a source term string with the appropriate OSV concept string using the term frequency– inverse document frequency method [8]. However, the Usagi matching engine is based on a bag-of-words model [9], which does not account for the semantic meanings of terms. This limitation affects its effectiveness in translating drug names from non-English-speaking countries.
A major challenge in international medical research is the comparison of drug prescriptions across countries. This difficulty arises from inconsistencies in drug names, dosages, forms, packaging, and administration methods [10]. Simple string comparisons often fail to achieve high mapping accuracy, leading researchers to explore semantics-based matching through sentence vector comparison as an alternative [11]. Additionally, the use of different measurement units across countries necessitates conversions and calculations, further complicating accurate drug comparisons. Evaluating similarity through simple embedding vector methods often falls short in identifying refined mapping candidates, thus requiring human verification of results. Automating this manual verification process with machine learning models could significantly enhance the efficiency of the mapping process.
The inference capability of large language models (LLMs) improves with an increase in the number of parameters, enhancing their ability to perform various linguistic tasks [12]. However, the volume of information an LLM can process at any given time is limited by the number of tokens it can handle, which complicates the selection of candidates from a large set. There, integrating an LLM with a retrieval-augmented generation (RAG) architecture [13], which functions like a search engine, can help the LLM generate sentences using information sourced by the search engine and consolidated within the token input limits. For instance, the recent integration of RAG with a few-shot learning algorithm enabled an LLM with fewer parameters to achieve performance levels comparable to those of a larger LLM [14].
In this study, we first investigated an architecture in which the RAG component initially extracted candidate drugs from an external vector database, based on the similarities of the drug-embedding vectors. These candidates were then embedded in a prompt that defined the LLM mapping evaluation criteria. The goal was to achieve higher accuracy than conventional systems, which rely solely on similarity evaluations when suggesting mapping candidates. We tested several LLMs to determine whether differences in their reasoning capabilities affected mapping accuracy.
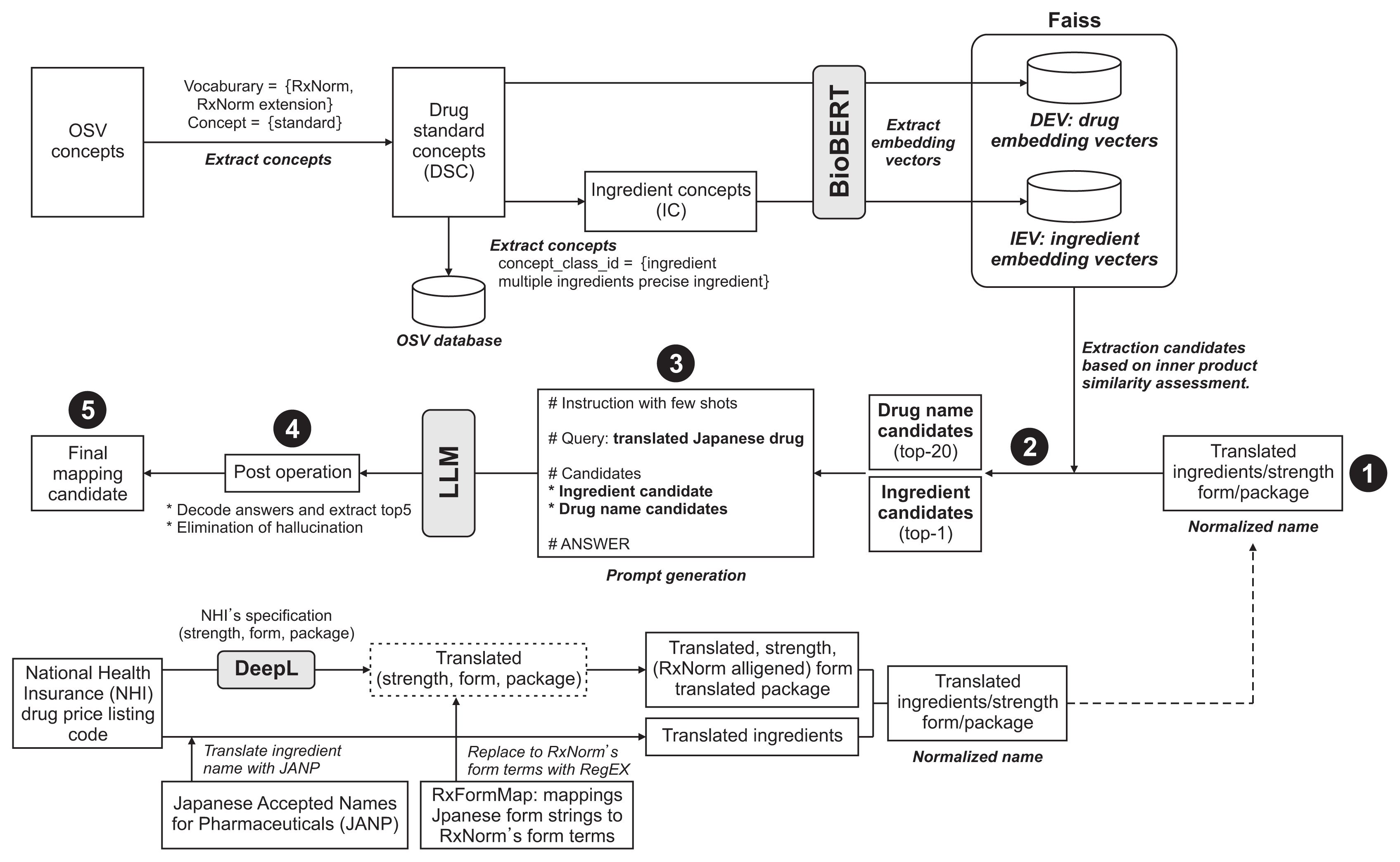
Currently, no open data sources in Japan offer detailed, structured information on drug components, forms, and dosages. To ensure that our results could be freely shared as open data, we decided against using any databases that impose license restrictions on reuse. Consequently, we utilized the National Health Insurance (NHI) drug price listing code (NHIDPLC) [15], which is published by the Ministry of Health, Labor and Welfare of Japan, as our data source for drug information in Japan. We translated Japanese drug names following the steps illustrated in Figure 1.
In Japan, medications are typically categorized into four types: PO (“naifukuyaku”), topical (“gaiyoyaku”), dental medicines, and injections. PO medicines, which are taken orally, are absorbed through the digestive system. This category includes tablets, capsules, and liquids that are used to treat infections, manage chronic diseases, and provide pain relief. Topical medicines are applied directly to the skin, eyes, or mucous membranes. This group comprises creams, ointments, lotions, and patches designed to treat skin conditions, muscle pain, and inflammation.
To translate drug ingredients, we used the Japanese Accepted Names for Pharmaceuticals (JANP) [16] database, which provides Japanese-English translations of drug ingredients. We developed a mapping table (RxFormMap) that uses regular expressions to deduce dosage form information from brand names and aligns Japanese dosage form names with those in RxNorm. Dosage forms and doses from NHIDPLC were translated using DeepL. The translated ingredients, dosages, and forms were then merged to create normalized terms for mapping.
Using ATHENA [17], we extracted the standard concepts from the RxNorm and RxNorm Extension vocabularies within the OSV version 5.0, dated August 31, 2023. The Semantic Clinical Drug class provided the drug standard concepts (DSCs). Concepts identified by the concept_class_id attributes of ingredient, multiple ingredients, or precise ingredient were designated as ingredient concepts. The concept_ name attribute was transformed into an embedding vector using the BioBERT language model [18], which uses the WordPiece tokenizer [19] to segment the input text. This tokenizer breaks down words into subwords, which helps in effectively managing terms that are not in the vocabulary. Each token is then mapped to an embedding vector that captures meaning and context by summing three components: token, position, and segment embeddings. We constructed a vector database containing the drug-embedding vectors (DEVs) and ingredient-embedding vectors (IEVs) for clinical drugs and ingredients using BioBERT. To facilitate rapid searches for similar vectors, we employed Faiss [20] as the vector database engine, and an index of exact similarity (IndexFlatIP: exact search for inner product [21]) was created using the inner product.
The BioBERT, Mistral 7b [22], Mixtral 8x7b [23], and GPT-3.5-turbo-0301 language models were tested as LLMs for integration into the LLM + RAG model. BioBERT demonstrated high-performance in biomedical natural language processing, leveraging its training on PubMed data [24]. We used BioBERT as a sentence vector extractor for drug names; this served as a baseline for comparing the performance of LLM + RAG models with those of traditional sentence vector similarity-based methods. Mistral 7b and Mixtral 8x7b were used as open-weight model LLMs for performance evaluation by comparing the evolution of the LLMs. Mixtral 8x7b is an open-weight model with a mixture-of-experts architecture [25] based on Mistral 7b, which effectively has a scale of 45 billion parameters [22]. At present, GPT-4 [26] is OpenAI’s state-of-the-art model, whereas GPT-3.5-turbo-1106 is a snapshot of OpenAI’s GPT-3.5 [27] model as of November 2023, which was selected as a high-performance yet cost-effective commercial LLM.
We developed a system that maps Japanese drugs to OSV drugs. The names of the Japanese drugs targeted for mapping were entered as normalized English strings (normalized names) following the process outlined in “1 Datasets” (➀ in Figure 1). Initially, we utilized BioBERT to generate the embedding vector for each Normalized Name. Subsequently, as part of the RAG process, we identified mapping candidates by selecting the top 20 clinical drugs based on the highest embedding vector similarities from the Faiss-hosted DEV database, along with the top single ingredient from the IEV database (➁ in Figure 1). These mapping candidates were then incorporated into the prompt using the LangChain prompt template feature [28] (➂ in Figure 1, Supplement A).
The prompt template is divided into sections titled “Instruction,” “Examples,” and “Question,” which present mapping targets, mapping candidates, and answers, respectively. The “Instruction” section includes detailed information on various aspects such as query dissection, examination of mapping options, selection criteria, maintaining accuracy and relevance, response formatting, clarification of dosage forms, handling of different dosage units, addressing partial information in query responses, managing additional ingredients in options, and treating variants of ingredient, brand, and generic names. It also covers the exclusion of unlisted information and the importance of match accuracy. Two mapping examples are provided in the few-shot algorithm instructions [12]. The dynamically generated prompt is used to query the LLM, and during post-processing, the top five mapping candidates are extracted from the LLM responses (➃ in Figure 1). To prevent LLM hallucinations, all candidate strings were verified against the DSC database; those not found were excluded. If the LLM produced fewer than five responses, “N/A” (not applicable) was added to the list until five answers were compiled, resulting in the final list of mapping candidates (➄ in Figure 1). This procedure was repeated with several LLMs. In comparing mapping performances, the top five embedding vector similarities from the BioBERT model were used as the baseline.
Two pharmacists with experience in university hospitals developed the ground truth dataset (Supplement B). This dataset includes, from the first column to the last, item names in Japanese, item names in English, and the drugs used to verify the mappings. The details provided for the drugs to be mapped encompassed the category (PO or topical), the Japanese drug code, the ingredient name, the standard dose, the product name, the manufacturer name, the original drug name, and the ingredient standard, all of which are in Japanese. The English translations for the ingredient and drug names are listed under the “Ingredient_DeepL” and “Drug-Name_DeepL” fields, respectively. The latter incorporates common descriptors of dosage forms and other parameters to generate the final English drug name used for mapping, referred to as the “normalized name.” The top 20 candidate clinical drug names identified by RAG were then presented, followed by the candidate ingredients. Evaluators compared each “Normalized Name” with the suggested drug name to determine matches for the ingredient (I), dosage (D), and dosage form (F). A complete match across these categories was labeled “IDF”; a match of only the ingredient and dosage was labeled “IF”. The absence of any match was labeled “NG”.
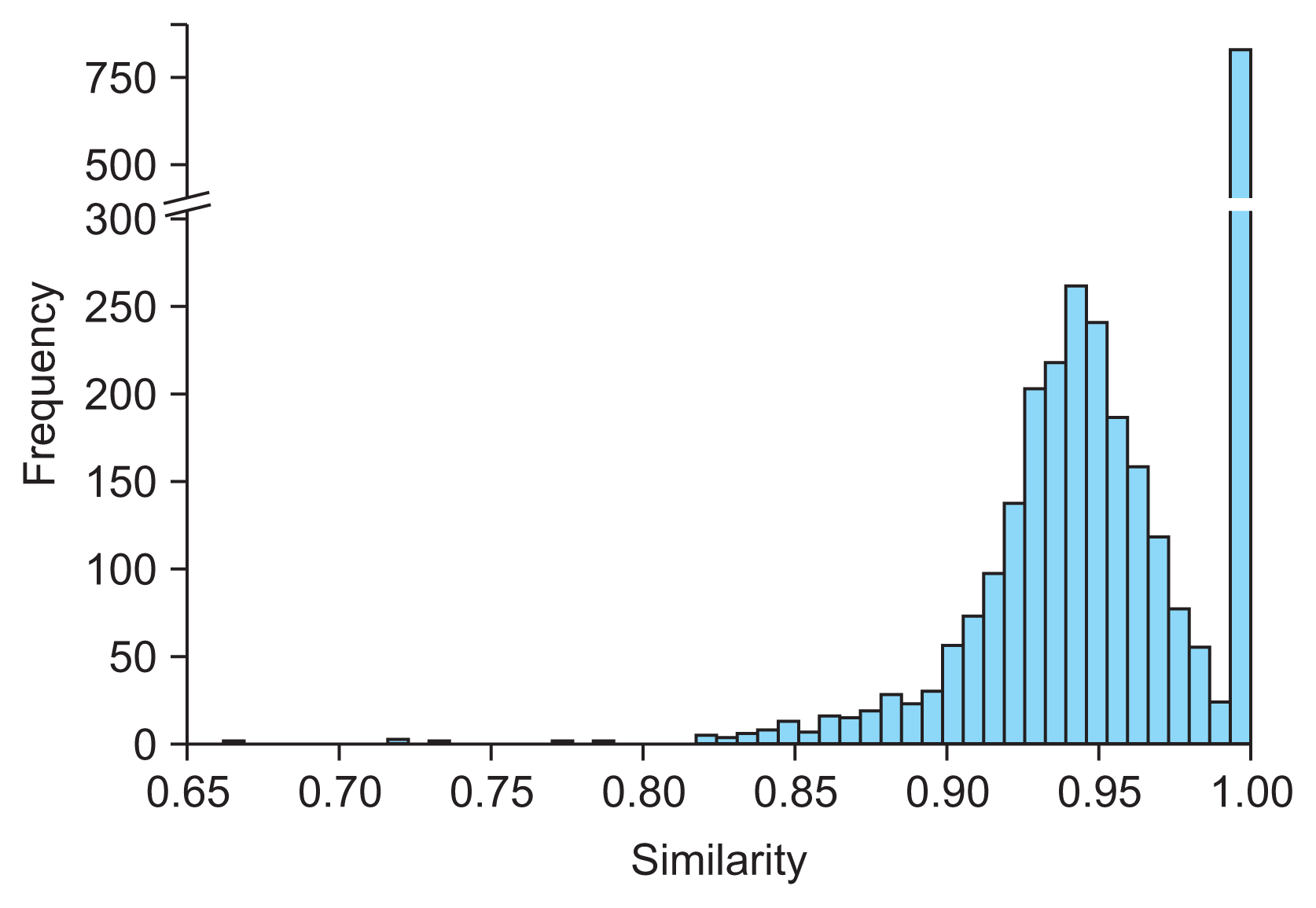
The BioBERT model, unlike LLM + RAG, is unable to select a final candidate from a group of potential options. Initially, we identified embedding vectors that surpassed a specific similarity threshold as final candidates. We hypothesized that ingredient names would show less variation in their string representations compared to drug names, thereby increasing the likelihood of achieving near-exact matches with very high similarity scores. We believed that using such similarity scores as thresholds would allow us to extract highly accurate candidates. To establish each threshold, we compared the JANP ingredient names with those from DEV, focusing on the similarities of their embedding vectors. We collected the highest similarity scores returned and visualized their distribution using a histogram (Figure 2). This analysis revealed a bimodal distribution with a distinct boundary at a similarity score of 0.985. Consequently, we adopted 0.985 as the threshold for BioBERT to identify candidate ingredients and map clinical drug names.
We sampled 100 oral and injectable medications during the mapping process. This selection was based on ingredient similarity and intentionally excluded products such as Chinese herbal medicines and pain relief patches unique to Japan, due to the absence of ingredient information and/or lack of identifiable clues, which could decrease the accuracy of correspondence mapping in the OSV. Relevance scores were assigned as follows: a score of 8 for matching ingredient, dose, and form (IDF); a score of 4 for matching ingredient and either dose or form (ID or IF); a score of 2 for matching ingredient only (I); and a score of 1 for none (N/A). The LLM matching results ranked the first as 1.0, decreasing by 0.2 points per rank down to 0.1 for N/A. The ranx Python library [29] was utilized to evaluate the results, employing various metrics including the number of hits (Hits), hit rate (Hit rate), r-precision (R-Prec), rank-biased precision (RBP), mean reciprocal rank (MRR), mean average precision (MAP), and normalized discounted cumulative gain (NDCG) [30]. RBP was evaluated assuming a highly impatient user, setting the persistence value at 0.5 [31]. Hits represent the average number of appropriate OSV drugs in the top five mapping candidates. The OSV included at least one entry for each class (IDF, ID, IF, and I); theoretically, one to more than four candidates could be obtained. The hit rates indicate the likelihood that appropriate OSV drugs were among the mapping candidates. R-Prec evaluates the number of drugs that were deemed correct by the ground truth, reflecting the alignment between LLM and human judgment. MAP assesses the average precision of a model in identifying relevant candidates, while MRR measures the model’s ability to quickly identify appropriate candidates. NDCG assesses the relevance and ranking of all information; higher scores signify better performance.
We employed the Fisher randomization test to assess various LLM performance metrics for several reasons. First, this non-parametric method does not assume a specific underlying data distribution, which was particularly suitable given that the performance metrics of different models were unlikely to adhere to a normal distribution. Secondly, the Fisher test proves invaluable when dealing with limited sample sizes. In our case, the evaluation dataset was relatively small and stratified.
The NHI database included 14,711 brand-name drugs, from which 5,438 unique semantic clinical drugs were identified following the normalization of components, dosages, and forms. A total of 2,638 unique components were identified (Table 1). In our analysis of 200 sampled mappings to OSV concepts for ground truth verification, we observed 137 mappings in the IDF class (68.5%), 95 in the IF class (47.5%), 92 in the ID class (46%), and 98 in the I class (49%).
Table 2 presents the evaluation results for drug mapping, based on a comparison of the LLMs using the Fisher randomization test. The results are stratified into groups: PO medicines, injections, and their combination. Below, we first discuss the overall results, followed by the specific findings for PO medicines and injections.
Drug mapping results of the BioBERT, Mistral 7b, Mixtral 8x7b, and GPT-3.5 models
The OSV contained at least one entry for each class (IDF, ID, IF, and I); therefore, the Hits metric could range from one to over four. This metric revealed that all LLMs significantly outperformed BioBERT. The Hit Rate metric represents the likelihood that the appropriate OSV drugs were included among the mapping candidates. Compared to the baseline Hit Rate of 63.82% for BioBERT, the rates for both the Mixtral 8x7b (94.47%) and GPT-3.5 (90.45%) models exceeded 90%, with the differences being statistically significant. Notably, Mixtral 8x7b at 94.47% was significantly more effective than Mistral7b at 82.91%, indicating enhanced performance of the LLM model. The R-Prec metric assesses the alignment of LLM judgments with human judgments. All LLMs showed significant improvements over the BioBERT baseline of 23.37%, with Mixtral 8x7b demonstrating a twofold increase at 49.76%.
The MRR and MAP metrics demonstrated that the LLMs were significantly more accurate than the baseline models, with scores of 0.67–0.77 and 0.42–0.56 compared to 0.51 and 0.23, respectively. The differences in MAP were particularly notable. The Mixtral 8x7b’s MAP performance was double that of the baseline model (0.56), indicating that higher-rank LLM mappings are more accurate; more correct mappings achieve higher positions. This reduces the effort required by evaluators to assess the final mapping results. The NDCG scores for all LLMs (0.49, 0.61, 0.56) significantly surpassed that of the baseline (0.38), indicating an overall improvement in mapping quality. Generally, the injection data were not as accurate as the PO medicine data; however, across all metrics, the LLMs significantly outperformed the baseline. The lower Hit and Hit Rate scores for injections suggest that appropriate candidates may not have been effectively extracted during the RAG stage.
Achieving high accuracy in mapping drug names across different languages and controlled vocabularies poses a significant challenge for approaches based on string comparison. In contrast, methods that match drugs through the evaluation of embedding vector similarity have proven to be somewhat effective [32]. However, in the field of drug mapping, the vast array of dose and dosage form combinations complicates the process of identifying the most appropriate mapping target from a multitude of candidates. This study integrated various LLMs with an RAG and demonstrated a notable improvement in matching accuracy using this combined approach. One reason for this enhanced accuracy is the ability of LLMs to filter out options that fail to meet the criteria specified in the prompt, from a range of dosage forms and combinations, ultimately selecting the top five candidates based on their informational richness. Additionally, LLMs have successfully performed tasks beyond mere string-based comparisons, such as recognizing “faropenem sodium hydrate 0.1 mg/mg” and converting the units to match the corresponding Japanese listing of “faropenem sodium hydrate 100 mg/g.”
Based on the above results, certain conclusions can be drawn. Unlike traditional deep learning models that depend solely on pre-trained knowledge and are limited to fixed tasks, the LLM + RAG model dynamically integrates new external knowledge and uses prompts to define tasks flexibly. Consequently, this approach can significantly enhance the accuracy of concept mapping tasks.
In the ground truth, the number of OSV concept classes matched by the models exhibited the following order: I > IDF > IF > ID. The selection process, which involved choosing from both the DEV top 20 list and consistently including the top IEV, along with a preference for longer string lengths in IDF embedding vector evaluations, may have contributed to the lower rankings of IF and ID within the top 20 results. Approximately 1% of all pharmaceuticals did not match any concept class during ground truthing, and 2% failed to match at the ingredient level. These mismatch rates seem to be within an acceptable range, considering that some medications are exclusively marketed in either Japan or the United States.
Our research focused exclusively on exploring the potential of the LLM + RAG models to enhance performance in mapping tasks. As a result, we did not extensively pursue enhancements to LLM prompts or explore LLMs with superior inference capabilities, including commercial LLMs known for their higher accuracy in task execution. Future studies could yield more precise outcomes by improving these aspects of the models. It would also be beneficial to expand the model tasks to include the integration of Japanese medication transaction data into the common data model of the OHDSI. We initiated mapping at the clinical drug and ingredient levels rather than at the branded drug level due to the unique challenges posed by Japanese drug names. In Japan, many medications are marketed under various brand names and are often described solely in Japanese, making it difficult to directly map branded drugs.
The terms “ingredient level,” “clinical drug level,” and “brand drug level” are hierarchical drug concepts in Rx-Norm. The hierarchical drug concepts of RxNorm begin with the ingredient level, proceed to the clinical drug level, which includes information on dosage and form, and culminate at the brand drug level, which pertains to brand-name medications. RxNorm defines the relationships among these concepts. Identifying a drug at the clinical level facilitates the location of associated branded drugs, providing a more accurate comparison than relying solely on brand names. Therefore, we initially mapped at the clinical drug level before referencing the branded drug names.
Our decision to initially map at the clinical drug level was influenced by the differences in drug packaging practices between Japan and the United States. Mapping drugs at the ID or IF level necessitates additional information and its transformation, as well as the application of extract, transform, and load (ETL) logic to achieve accurate code mapping. This process requires data on dosages and unit conversions. Thus, columns containing unit conversion data and other coefficients need to be incorporated into existing mapping tables, which complicates the content generation process. LLM prompts are particularly useful in these scenarios.
In the current study, no drug knowledge database was used as a source for mapping; instead, drug names were compared solely on a string basis. If external drug knowledge databases included precise data on normalized ingredients, dosages, and formulations, the accuracy of mapping would likely improve further.
Acknowledgments
This research was supported by MEXT as ‘Developing a Research Data Ecosystem for the Promotion of Data-Driven Science.’
Supplementary Materials
Supplementary materials can be found via https://doi.org/10.4258/hir.2024.30.4.355.
References
1. Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform. 2015; 216:574–8.
2. Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, et al. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med. 2010; 153(9):600–6.
https://doi.org/10.7326/0003-4819-153-9-201011020-00010.

3. Reich C, Ostropolets A, Ryan P, Rijnbeek P, Schuemie M, Davydov A, et al. OHDSI standardized vocabularies-a large-scale centralized reference ontology for international data harmonization. J Am Med Inform Assoc. 2024; 31(3):583–90.
https://doi.org/10.1093/jamia/ocad247.

4. Wang L, Zhang Y, Jiang M, Wang J, Dong J, Liu Y, et al. Toward a normalized clinical drug knowledge base in China-applying the RxNorm model to Chinese clinical drugs. J Am Med Inform Assoc. 2018; 25(7):809–18.
https://doi.org/10.1093/jamia/ocy020.

5. Henke E, Zoch M, Kallfelz M, Ruhnke T, Leutner LA, Spoden M, et al. Assessing the use of German claims data vocabularies for research in the observational medical outcomes partnership common data model: development and evaluation study. JMIR Med Inform. 2023; 11:e47959.
https://doi.org/10.2196/47959.

6. Maier C, Lang L, Storf H, Vormstein P, Bieber R, Bernarding J, et al. Towards implementation of OMOP in a German University Hospital Consortium. Appl Clin Inform. 2018; 9(1):54–61.
https://doi.org/10.1055/s-0037-1617452.

7. Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011; 18(4):441–8.
https://doi.org/10.1136/amiajnl-2011-000116.

8. de Groot R, Puttmann DP, Fleuren LM, Thoral PJ, Elbers PW, de Keizer NF, et al. Determining and assessing characteristics of data element names impacting the performance of annotation using Usagi. Int J Med Inform. 2023. Oct. 178:105200.
https://doi.org/10.1016/j.ijmedinf.2023.105200.

9. Zhang Y, Jin R, Zhou ZH. Understanding bag-of-words model: a statistical framework. Int J Mach Learn Cybern. 2010; 1(1):43–52.
https://doi.org/10.1007/s13042-010-0001-0.

10. Butler J, Zand M. Similarity mapping of national drug code formulary systems between nations [Internet]. Durham (NC): Research Square;2022. [cited at 2024 Jun 6]. Available from: https://doi.org/10.21203/rs.3.rs-1858694/v1.

11. Zhang Y, Guo L, Du C, Wang Y, Huang D. Extraction of English drug names based on Bert-CNN mode. J Inf Hiding Multimed Signal Process. 2020; 11(2):70–8.
12. Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al. Language models are few-shot learners. Adv Neural Inf Process Syst. 2020; 33:1877–901.
https://doi.org/10.48550/arXiv.2005.14165.

13. Gao Y, Xiong Y, Gao X, Jia K, Pan J, Bi Y, et al. Retrieval-augmented generation for large language models: a survey [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2005.14165.
14. Izacard G, Lewis P, Lomeli M, Hosseini L, Petroni F, Schick T, et al. Few-shot learning with retrieval augmented language models [Internet]. Ithaca (NY): arXiv.org;2022. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2208.03299.
15. Ministry of Health, Labour and Welfare. National Health Insurance (NHI) drug price listing code 2024 [Internet]. Tokyo, Japan: Ministry of Health, Labour and Welfare;c2024. [cited at 2024 Jun 6]. Available from: https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/0000078916.html.
16. National Institute of Health Sciences. Japanese accepted names for pharmaceuticals [Internet]. Tokyo, Japan: National Institute of Health Sciences;c2024. [cited at 2024 Jun 6]. Available from: https://jpdb.nihs.go.jp/jan.
17. Odysseus Data Services. ATHENA: OHDSI Vocabularies Repository [Internet]. [place unknown]: OHDSI;c2024. [cited at 2024 Jun 6]. Available from: https://athena.ohdsi.org/.
18. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioB-ERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020; 36(4):1234–40.
https://doi.org/10.1093/bioinformatics/btz682.

19. Wu Y. Google’s neural machine translation system: bridging the gap between human and machine translation [Internet]. Ithaca (NY): arXiv.org;2016. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/1609.08144.
20. Douze M, Guzhva A, Deng C, Johnson J, Szilvasy G, Mazare PE, et al. The Faiss library [Internet]. Ithaca (NY): arXiv.org;2024. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2401.08281.
21. Lomeli M. Faiss indexes [Internet]. San Francisco (CA): GitHub;2023. [cited at 2024 Jun 6]. Available from: https://github.com/facebookresearch/faiss/wiki/Faissindexes.
22. Jiang AQ, Sablayrolles A, Mensch A, Bamford C, Chaplot DS, de las Casas D, et al. Mistral 7B [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2310.06825.
23. Tunstall L, Schmid P, Sanseviero O, Cuenca P, Dehaene O, von Werra L, et al. Welcome Mixtral: a SOTA mixture of experts on Hugging Face [Internet]. Brooklyn (NY): Hugging Face;2023. [cited at 2024 Jun 6]. Available from: https://huggingface.co/blog/mixtral.
24. Ji Z, Wei Q, Xu H. BERT-based ranking for biomedical entity normalization. AMIA Jt Summits Transl Sci Proc. 2020; 2020:269–77.
25. Shazeer N, Mirhoseini A, Maziarz K, Davis A, Le Q, Hinton G, et al. Outrageously large neural networks: the sparsely-gated mixture-of-experts layer [Internet]. Ithaca (NY): arXiv.org;2017. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/1701.06538.
26. Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, Aleman FL, et al. GPT-4 technical report [Internet]. Ithaca (NY): arXiv.org;2023. [cited at 2024 Jun 6]. Available from: https://arxiv.org/abs/2303.08774v1.
27. Wu T, He S, Liu J, Sun S, Liu K, Han QL, et al. A brief overview of ChatGPT: the history, status quo and potential future development. IEEE/CAA J Automatica Sinica. 2023; 10(5):1122–36.
https://doi.org/10.1109/JAS.2023.123618.

28. GitHub. LangChain [Internet]. San Francisco (CA): GitHub;2022. [cited at 2024 Jun 6]. Available from: https://github.com/langchain-ai/langchain.
29. Bassani E. ranx: a blazing-fast python library for ranking evaluation and comparison. European Conference on Information Retrieval. Cham, Switzerland: Springer;2022. p. 259–64.
https://doi.org/10.1007/978-3-030-99739-7_30.

30. Jarvelin K, Kekalainen J. Cumulated gain-based evaluation of IR techniques. ACM Trans Inf Syst. 2002; 20(4):422–46.
https://doi.org/10.1145/582415.582418.

31. Moffat A, Zobel J. Rank-biased precision for measurement of retrieval effectiveness. ACM Trans Inf Syst. 2008; 27(1):1–27.
https://doi.org/10.1145/1416950.1416952.

32. Chen Y, Hu D, Li M, Duan H, Lu X. Automatic SNOMED CT coding of Chinese clinical terms via attention-based semantic matching. Int J Med Inform. 2022; 159:104676.
https://doi.org/10.1016/j.ijmedinf.2021.104676.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download