1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence
and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68(6):394–424. DOI:
10.3322/caac.21492. PMID:
30207593.

5. Gordan JD, Kennedy EB, Abou-Alfa GK, Beg MS, Brower ST, Gade TP, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO
guideline. J Clin Oncol. 2020; 38(36):4317–4345. DOI:
10.1200/JCO.20.02672. PMID:
33197225.
6. Doycheva I, Thuluvath PJ. Systemic therapy for advanced hepatocellular carcinoma: an update
of a rapidly evolving field. J Clin Exp Hepatol. 2019; 9(5):588–596. DOI:
10.1016/j.jceh.2019.07.012. PMID:
31695249. PMCID:
PMC6823698.

8. Burt AD, Ferrell LD, Hübscher SG. MacSween's pathology of the liver. Amsterdam: Elsevier Health Sciences;2022.
9. Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, et al. The 2019 WHO classification of tumours of the digestive
system. Histopathology. 2020; 76(2):182–188. DOI:
10.1111/his.13975. PMID:
31433515. PMCID:
PMC7003895.

11. Martins-Filho SN, Paiva C, Azevedo RS, Alves VAF. Histological grading of hepatocellular carcinoma: a systematic
review of literature. Front Med. 2017; 4:193. DOI:
10.3389/fmed.2017.00193. PMID:
29209611. PMCID:
PMC5701623.
13. The Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of
hepatocellular carcinoma. Cell. 2017; 169(7):1327–1341.E23.
14. Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc JF, et al. Histological subtypes of hepatocellular carcinoma are related to
gene mutations and molecular tumour classification. J Hepatol. 2017; 67(4):727–738. DOI:
10.1016/j.jhep.2017.05.014. PMID:
28532995.

15. Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma
derived from hepatic progenitor cells. Nat Med. 2006; 12(4):410–416. DOI:
10.1038/nm1377. PMID:
16532004.

16. Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene
alterations and to new therapeutic targets. Hepatology. 2007; 45(1):42–52. DOI:
10.1002/hep.21467. PMID:
17187432.

17. Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of
hepatocellular carcinoma. Cancer Res. 2008; 68(16):6779–6788. DOI:
10.1158/0008-5472.CAN-08-0742. PMID:
18701503. PMCID:
PMC2587454.

18. Hoshida Y, Nijman SMB, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular
subclasses of human hepatocellular carcinoma. Cancer Res. 2009; 69(18):7385–7392. DOI:
10.1158/0008-5472.CAN-09-1089. PMID:
19723656. PMCID:
PMC3549578.

19. Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, et al. Identification of an immune-specific class of hepatocellular
carcinoma, based on molecular features. Gastroenterology. 2017; 153(3):812–826. DOI:
10.1053/j.gastro.2017.06.007. PMID:
28624577.

20. Audard V, Grimber G, Elie C, Radenen B, Audebourg A, Letourneur F, et al. Cholestasis is a marker for hepatocellular carcinomas displaying
β-catenin mutations. J Pathol. 2007; 212(3):345–352. DOI:
10.1002/path.2169. PMID:
17487939.

21. Salomao M, Yu WM, Brown RS Jr, Emond JC, Lefkowitch JH. Steatohepatitic hepatocellular carcinoma (SH-HCC): a distinctive
histological variant of HCC in hepatitis C virus-related cirrhosis with
associated NAFLD/NASH. Am J Surg Pathol. 2010; 34(11):1630–1636. DOI:
10.1097/PAS.0b013e3181f31caa. PMID:
20975341.
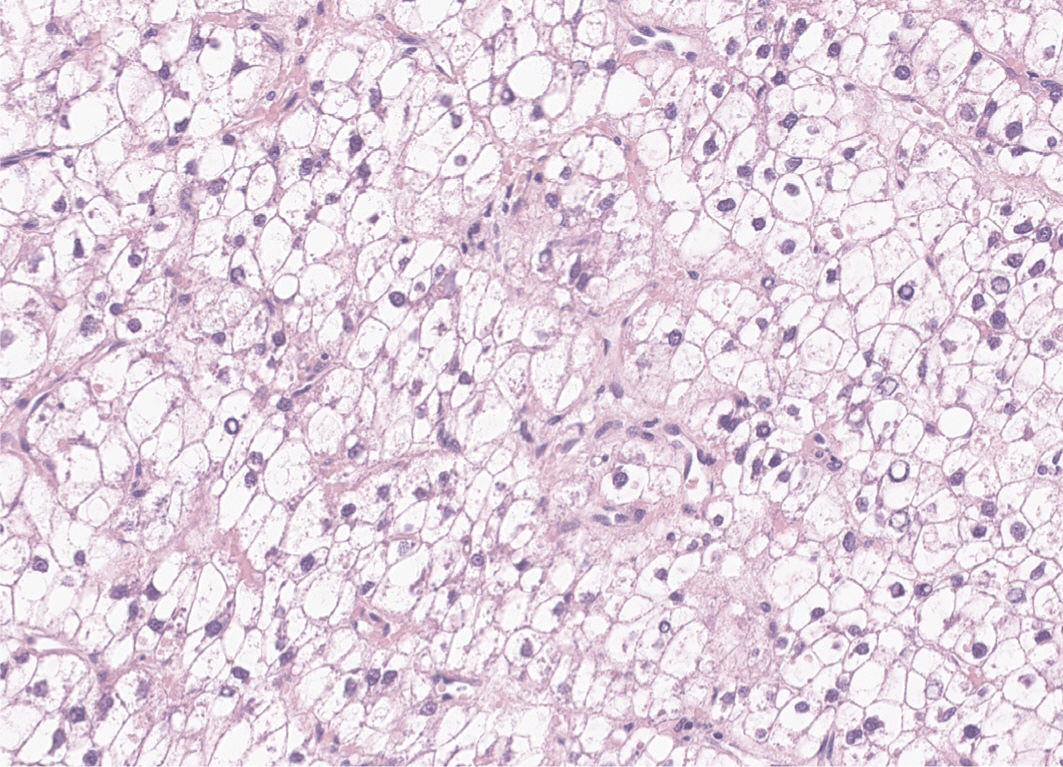
22. Bannasch P, Ribback S, Su Q, Mayer D. Clear cell hepatocellular carcinoma: origin, metabolic traits and
fate of glycogenotic clear and ground glass cells. Hepatobiliary Pancreatic Dis Int. 2017; 16(6):570–594. DOI:
10.1016/S1499-3872(17)60071-7. PMID:
29291777.

23. Li T, Fan J, Qin LX, Zhou J, Sun HC, Qiu SJ, et al. Risk factors, prognosis, and management of early and late
intrahepatic recurrence after resection of primary clear cell carcinoma of
the liver. Ann Surg Oncol. 2011; 18(7):1955–1963. DOI:
10.1245/s10434-010-1540-z. PMID:
21240562.

24. Lee JH, Shin DH, Park WY, Shin N, Kim A, Lee HJ, et al. IDH1 R132C mutation is detected in clear cell hepatocellular
carcinoma by pyrosequencing. World J Surg Oncol. 2017; 15(1):82. DOI:
10.1186/s12957-017-1144-1. PMID:
28403884. PMCID:
PMC5389153.
25. Jeon Y, Benedict M, Taddei T, Jain D, Zhang X. Macrotrabecular hepatocellular carcinoma: an aggressive subtype
of hepatocellular carcinoma. Am J Surg Pathol. 2019; 43(7):943–948. DOI:
10.1097/PAS.0000000000001289. PMID:
31135484.
26. Kumar D, Hafez O, Jain D, Zhang X. Can primary hepatocellular carcinoma histomorphology predict
extrahepatic metastasis? Hum Pathol. 2021; 113:39–46. DOI:
10.1016/j.humpath.2021.04.008. PMID:
33905775.

27. Renne SL, Woo HY, Allegra S, Rudini N, Yano H, Donadon M, et al. Vessels encapsulating tumor clusters (VETC) is a powerful
predictor of aggressive hepatocellular carcinoma. Hepatology. 2020; 71(1):183–195. DOI:
10.1002/hep.30814. PMID:
31206715.

28. Fang JH, Xu L, Shang LR, Pan CZ, Ding J, Tang YQ, et al. Vessels that encapsulate tumor clusters (VETC) pattern is a
predictor of sorafenib benefit in patients with hepatocellular
carcinoma. Hepatology. 2019; 70(3):824–839. DOI:
10.1002/hep.30366. PMID:
30506570.

29. Kurogi M, Nakashima O, Miyaaki H, Fujimoto M, Kojiro M. Clinicopathological study of scirrhous hepatocellular
carcinoma. J Gastroenterol Hepatol. 2006; 21(9):1470–1477. DOI:
10.1111/j.1440-1746.2006.04372.x. PMID:
16911695.
30. Seok JY, Na DC, Woo HG, Roncalli M, Kwon SM, Yoo JE, et al. A fibrous stromal component in hepatocellular carcinoma reveals a
cholangiocarcinoma-like gene expression trait and epithelial-mesenchymal
transition. Hepatology. 2012; 55(6):1776–1786. DOI:
10.1002/hep.25570. PMID:
22234953.

31. Matsuura S, Aishima S, Taguchi K, Asayama Y, Terashi T, Honda H, et al. 'Scirrhous' type hepatocellular carcinomas: a
special reference to expression of cytokeratin 7 and hepatocyte paraffin
1. Histopathology. 2005; 47(4):382–390. DOI:
10.1111/j.1365-2559.2005.02230.x. PMID:
16178893.

32. Chan AWH, Tong JHM, Pan Y, Chan SL, Wong GLH, Wong VWS, et al. Lymphoepithelioma-like hepatocellular carcinoma: an uncommon
variant of hepatocellular carcinoma with favorable outcome. Am J Surg Pathol. 2015; 39(3):304–312. DOI:
10.1097/PAS.0000000000000376. PMID:
25675010.
33. Calderaro J, Rousseau B, Amaddeo G, Mercey M, Charpy C, Costentin C, et al. Programmed death ligand 1 expression in hepatocellular carcinoma:
relationship with clinical and pathological features. Hepatology. 2016; 64(6):2038–2046. DOI:
10.1002/hep.28710. PMID:
27359084.

34. Nishida N, Sakai K, Morita M, Aoki T, Takita M, Hagiwara S, et al. Association between genetic and immunological background of
hepatocellular carcinoma and expression of programmed cell
death-1. Liver Cancer. 2020; 9(4):426–439. DOI:
10.1159/000506352. PMID:
32999869. PMCID:
PMC7506256.

35. Chan AWH, Zhang Z, Chong CCN, Tin EKY, Chow C, Wong N. Genomic landscape of lymphoepithelioma-like hepatocellular
carcinoma. J Pathol. 2019; 249(2):166–172. DOI:
10.1002/path.5313. PMID:
31168847.
37. El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular
carcinoma? A US population-based study. Hepatology. 2004; 39(3):798–803. DOI:
10.1002/hep.20096. PMID:
14999699.

38. Graham RP, Yeh MM, Lam-Himlin D, Roberts LR, Terracciano L, Cruise MW, et al. Molecular testing for the clinical diagnosis of fibrolamellar
carcinoma. Mod Pathol. 2018; 31(1):141–149. DOI:
10.1038/modpathol.2017.103. PMID:
28862261. PMCID:
PMC5758901.

39. Gougelet A, Torre C, Veber P, Sartor C, Bachelot L, Denechaud PD, et al. T-cell factor 4 and β-catenin chromatin occupancies
pattern zonal liver metabolism in mice. Hepatology. 2014; 59(6):2344–2357. DOI:
10.1002/hep.26924. PMID:
24214913.

41. Ueno A, Masugi Y, Yamazaki K, Komuta M, Effendi K, Tanami Y, et al. OATP1B3 expression is strongly associated with
Wnt/β-catenin signalling and represents the transporter of gadoxetic
acid in hepatocellular carcinoma. J Hepatol. 2014; 61(5):1080–1087. DOI:
10.1016/j.jhep.2014.06.008. PMID:
24946283.

42. Torbenson M, McCabe CE, O'Brien DR, Yin J, Bainter T, Tran NH, et al. Morphological heterogeneity in beta-catenin–mutated
hepatocellular carcinomas: implications for tumor molecular
classification. Hum Pathol. 2022; 119:15–27. DOI:
10.1016/j.humpath.2021.09.009. PMID:
34592239. PMCID:
PMC9258524.

43. Rhee H, Kim H, Park YN. Clinico-radio-pathological and molecular features of
hepatocellular carcinomas with Keratin 19 expression. Liver Cancer. 2020; 9(6):663–681. DOI:
10.1159/000510522. PMID:
33442539. PMCID:
PMC7768132.

44. Fang JH, Zhou HC, Zhang C, Shang LR, Zhang L, Xu J, et al. A novel vascular pattern promotes metastasis of hepatocellular
carcinoma in an epithelial–mesenchymal transition–independent
manner. Hepatology. 2015; 62(2):452–465. DOI:
10.1002/hep.27760. PMID:
25711742.

46. Wood LD, Heaphy CM, Daniel HDJ, Naini BV, Lassman CR, Arroyo MR, et al. Chromophobe hepatocellular carcinoma with abrupt anaplasia: a
proposal for a new subtype of hepatocellular carcinoma with unique
morphological and molecular features. Mod Pathol. 2013; 26(12):1586–1593. DOI:
10.1038/modpathol.2013.68. PMID:
23640129. PMCID:
PMC3974906.

48. Amano H, Itamoto T, Emoto K, Hino H, Asahara T, Shimamoto F. Granulocyte colony-stimulating factor-producing combined
hepatocellular/cholangiocellular carcinoma with sarcomatous
change. J Gastroenterol. 2005; 40(12):1158–1159. DOI:
10.1007/s00535-005-1715-8. PMID:
16378181.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download