Abstract
Background
Methods
Results
Supplementary Material
Supplemental Table S2.
Supplemental Fig. S1.
Supplemental Fig. S2.
Supplemental Fig. S3.
Notes
CONFLICTS OF INTEREST
Young Joo Park is an editor-in-chief and Sun Wook Cho is a deputy editor of the journal. But they were not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflicts of interest relevant to this article were reported.
Young Joo Park received fundings from DCGen Co. Ltd. (No. SNUH 0620204943 and 0620234950), but has no other conflicts of interest relevant to this article. Hye-Mi Choi, Woosung Chung, and Young Jun Chai, who are affiliated with DCGen Co. Ltd., had no influence on the outcomes of this study.
AUTHOR CONTRIBUTIONS
Conception or design: M.Y.O., H.M.C., W.C., Y.J.C., Y.J.P. Acquisition, analysis, or interpretation of data: M.Y.O., H.M.C. Drafting the work or revising: M.Y.O., H.M.C., W.C., Y.J.P. Final approval of the manuscript: M.Y.O., H.M.C., W.C., J.J., H.S., S.S.P., M.S., Y.H.K., S.W.C., Y.J.C., Y.J.P.
ACKNOWLEDGMENTS
REFERENCES
Fig. 1.
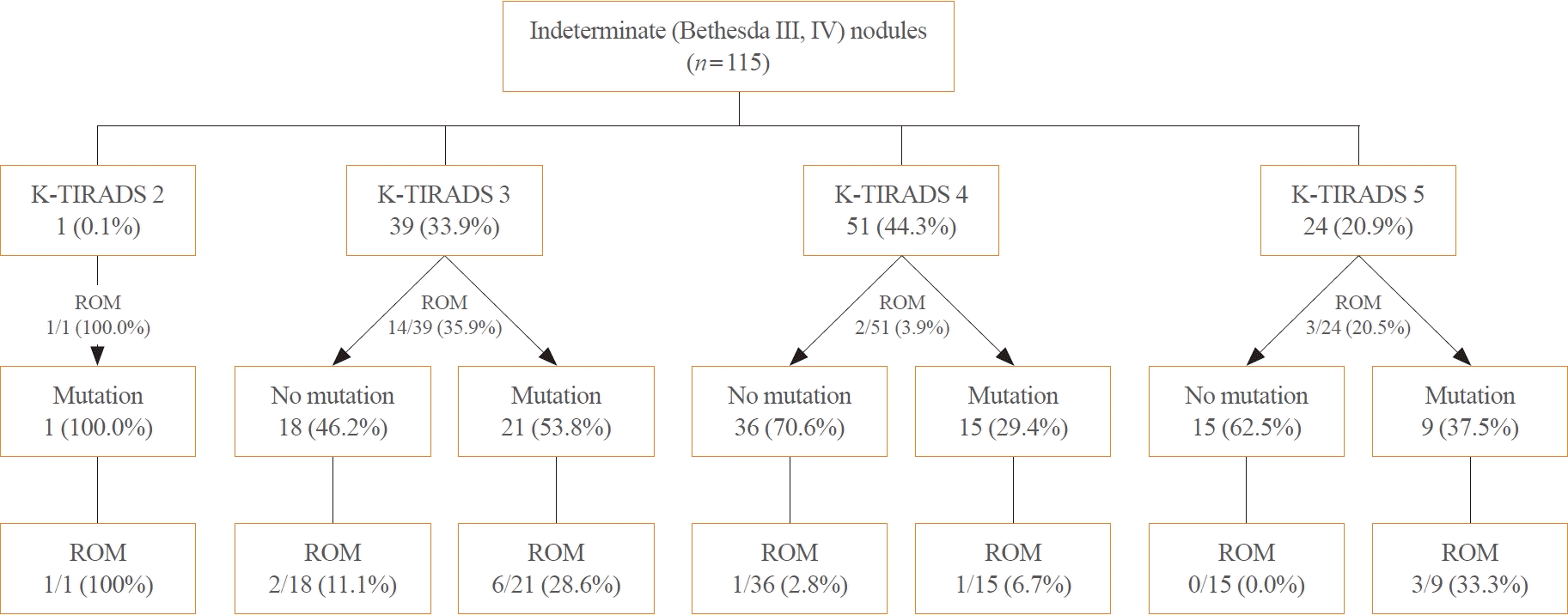
Fig. 2.
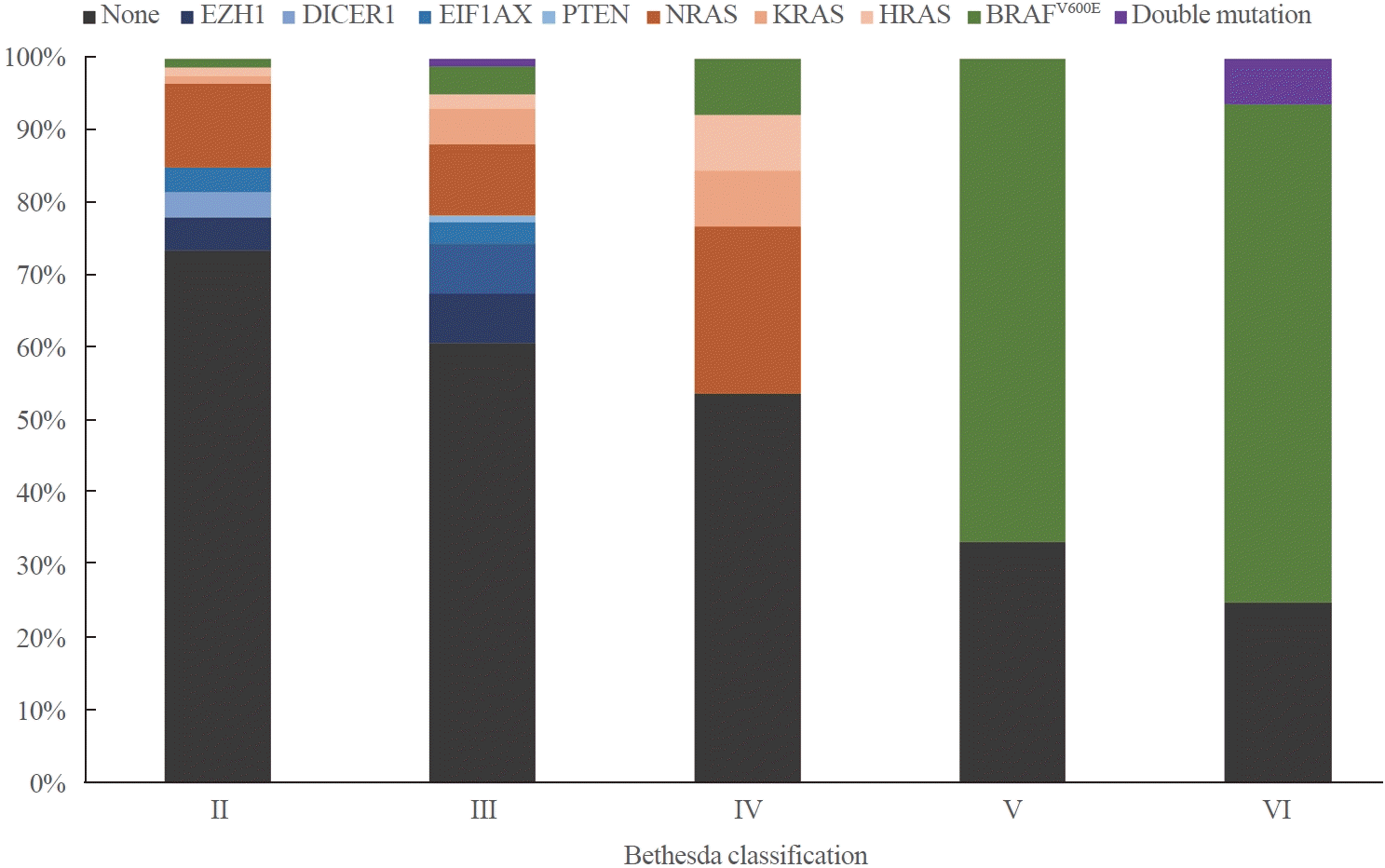
Table 1.
| Characteristic |
Before propensity score-matching |
After propensity score-matching |
||||||
|---|---|---|---|---|---|---|---|---|
| Control group (n=475) | Panel group (n=278) | SMD before matching | P value | Control group (n=278) | Panel group (n=278) | SMD after matching | P value | |
| Female sex | 401 (84.4) | 218 (78.4) | 0.155 | 0.038 | 223 (80.2) | 218 (78.4) | 0.044 | 0.601 |
| Age, yr | 54.6±14.3 | 54.0±13.4 | 0.044 | 0.565 | 53.6±14.4 | 54.0±13.4 | 0.036 | 0.733 |
| Nodule size, cm | 1.8±1.3 | 1.7±1.1 | 0.106 | 0.150 | 1.7±1.1 | 1.7±1.1 | 0.103 | 0.903 |
| K-TIRADS | 0.434 | –1.014 | ||||||
| 2 | 16 (3.4) | 1 (0.4) | <0.001 | 1 (0.4) | 1 (0.4) | 0.962 | ||
| 3 | 192 (40.4) | 73 (26.3) | 76 (27.3) | 73 (26.3) | ||||
| 4 | 166 (34.9) | 103 (37.1) | 106 (38.1) | 103 (37.1) | ||||
| 5 | 101 (21.3) | 101 (36.3) | 95 (34.2) | 101 (36.3) | ||||
| Bethesda classification | 0.105 | –0.045 | ||||||
| I | 27 (5.7) | 19 (6.8) | 0.085 | 20 (7.2) | 19 (6.8) | 0.859 | ||
| II | 192 (40.4) | 87 (31.3) | 88 (31.7) | 87 (31.3) | ||||
| III | 143 (30.1) | 102 (36.7) | 103 (37.1) | 102 (36.7) | ||||
| IV | 32 (6.7) | 13 (4.7) | 7 (2.5) | 13 (4.7) | ||||
| V | 17 (3.6) | 9 (3.2) | 9 (3.2) | 9 (3.2) | ||||
| VI | 64 (13.5) | 48 (17.3) | 51 (18.3) | 48 (17.3) | ||||
| Management | ||||||||
| Surgery | 221 (46.5) | 75 (27.0) | <0.001 | 146 (52.5) | 75 (27.0) | <0.001 | ||
| Surveillance | 254 (53.5) | 204 (73.4) | 132 (47.5) | 204 (73.4) | ||||
| Definite histopathologya | ||||||||
| Malignant | 103 (61.3) | 42 (72.4) | 0.175 | 74 (66.7) | 42 (72.4) | 0.588 | ||
| PTC | 83 | 34 | 60 | 34 | ||||
| DHGTC | - | 1 | - | 1 | ||||
| FTC | 10 | 5 | 8 | 5 | ||||
| OCA | - | 2 | - | 2 | ||||
| FT-UMP | 10 | - | 6 | - | ||||
| NIFTP | 8 (4.8) | 1 (1.7) | 6 (5.4) | 1 (1.7) | ||||
| Benign | 57 (33.9) | 15 (25.9) | 31 (27.9) | 15 (25.9) | ||||
| FA | 36 | 7 | 17 | 7 | ||||
| OA | 6 | 3 | 4 | 3 | ||||
| FND | 15 | 5 | 10 | 5 | ||||
Values are expressed as number (%) or mean±standard deviation. One-to-one propensity score-matching of sex, age, nodule size, K-TIRADS criteria, and Bethesda classification.
SMD, standardized mean difference; K-TIRADS, Korean Thyroid Imaging Reporting and Data System; PTC, papillary thyroid carcinoma; DHGTC, differentiated high-grade thyroid carcinoma; FTC, follicular thyroid carcinoma; OCA, oncocytic carcinoma of the thyroid; FT-UMP, follicular tumor of uncertain malignant potential; NIFTP, non-invasive follicular thyroid neoplasm with papillary-like nuclear features; FA, follicular adenoma; OA, oncocytic adenoma of the thyroid; FND, follicular nodular disease.
Table 2.
| Study |
Present study |
Commercial panels |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 11-Gene DNA panel |
7-Gene panel |
Afirma GEC |
Afirma GSC |
ThyroSeq v3 |
|||||
| Real world | Validation | Real world | Validation | Real world | Validation | Real world | |||
| Included nodules (included studies) | 278 | 115 | 956 (7) [20-25] | 265 (1) [26] | 793 (6) [27-32] | 190 (1) [7] | 1,976 (13 of 1 meta-analysis) [33] | 257 (1) [34] | 530 (6 of 1 meta-analysis) [35] |
| Bethesda category | All | III–IV | III–IV | III–IV | III–IV | III–IV | III–IV | III–IV | III–IV |
| Panel results | |||||||||
| PCR, % | 41.7 | 40.0 | 10–39a | 59 | 45–74a | 46 | 32.9 | 41 | 29–39a |
| BCR, % | 69.8 | 75.7 | 61–90a | 41 | 26–55a | 54 | 66.9 | 61 | 61–71a |
| Diagnostic performance | |||||||||
| No. of resected nodules | 75 | 27 | 956 | 265 | 360 | 190 | 582 | 257 | 530 |
| Sensitivity, % | 80.0 (68.2–88.9)b | 78.6 (49.2–95.3)b | 18–86a | 92 | 83–100a | 91 | 97 (91–100)c | 94 | 95 (59–86)c |
| Specificity, % | 53.3 (26.6–78.7)b | 45.5 (16.8–76.6)b | 82–99a | 52 | 8–24a | 68 | 88 (76–100)c | 82 | 50 (5–75)c |
| PPV, % | 88.1 (81.0–92.8)b | 64.7 (50.0–77.1)b | 19–88a | 47 | 75–100a | 47 | 65 (35–100)c | 66 | 70 (17–83)c |
| NPV, % | 38.1 (23.8–54.8)b | 62.5 (33.6–84.6)b | 56–97a | 93 | 14–57a | 96 | 99 (96–100)c | 97 | 92 (21–93)c |
Table 3.
Table 4.
|
Molecular group |
Mutated gene |
Pathology of resected nodules |
||||||
|---|---|---|---|---|---|---|---|---|
| Group | Total (n=278) | Resected (n=75, 27.0%) | Gene | Total (n=278) | Resected (n=75, 27.0%) | Malignant (n=43, 57.3%) | Benign (n=16, 21.3%) | Unknowna (n=16, 21.3%) |
| No mutation | 162 (58.3) | 30 (18.5) | Noneb | 162 (58.3) | 30 (18.5) | 12 (8 PTC, 2 FTC, 2 OCA) | 8 (3 FA, 1 OA, 4 FND) | 10 |
| NBNR | 32 (12.6) | 4 (12.5) | EZH1 | 12 (4.3) | 1 (8.3) | - | 1 (FA) | - |
| DICERc | 11 (4.0) | 1 (9.1) | - | 1 (FA) | - | |||
| EIF1AX | 7 (2.5) | 2 (28.6) | 1 (FTC) | 1 (FA) | - | |||
| PTEN | 2 (0.7) | - | - | - | - | |||
| RAS | 35 (12.6) | 9 (25.7) | NRAS | 24 (8.6) | 8 (33.3) | 4 (2 PTC, 1 FTC, 1 NIFTP) | 4 (1 FA, 1 OA, 1 FND) | 1 |
| KRASc | 7 (2.5) | - | - | - | - | |||
| HRAS | 4 (1.4) | 1 (25.0) | 1 (FTC) | - | - | |||
| BRAFV600E | 45 (16.2) | 28 (62.2) | BRAFV600Ec | 45 (16.2) | 28 (62.2) | 22 (21 PTC, 1 DHGTC) | - | 6 |
| Double mutation | 4 (1.4) | 4 (100.0) | BRAFV600E/PIK3CA | 1 (0.4) | 1 (100.0) | 1 (PTC) | - | - |
| BRAFV600E/TP53 | 1 (0.4) | 1 (100.0) | 1 (PTC) | - | ||||
| PIK3CA/TERT | 1 (0.4) | 1 (100.0) | 1 (PTC) | - | - | |||
| EZH1/TP53 | 1 (0.4) | 1 (100.0) | - | 1 (OA) | ||||
Values are expressed as number (%), unless otherwise specified.
NBNR, non-BRAF-non-RAS; EZH1, enhancer of zeste 1 polycomb repressive complex 2 subunit; DICER1, dicer 1, ribonuclease III; EIF1AX, eukaryotic translation initiation factor 1A X-linked; PTEN, phosphatase and tensin homolog; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; TP53, tumor protein p53; TERT, telomerase reverse transcriptase; PTC, papillary thyroid carcinoma; FTC, follicular thyroid carcinoma; OCA, oncocytic carcinoma of the thyroid; FA, follicular adenoma; OA, oncocytic adenoma of the thyroid; FND, follicular nodular disease; NIFTP, noninvasive follicular thyroid neoplasm with papillary-like nuclear features; DHGTC, differentiated high-grade thyroid carcinoma.
Table 5.
Table 6.
K-TIRADS, Korean Thyroid Imaging Reporting and Data System; ETE, extrathyroidal extension; LN, lymph node; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; VAF, variant allele frequency; RCND, right central node dissection; PTC, papillary thyroid carcinoma; TP53, tumor protein p53; BCND, bilateral central node dissection; TERT, telomerase reverse transcriptase; LCND, left central node dissection; EZH1, enhancer of zeste 1 polycomb repressive complex 2 subunit; OA, oncocytic adenoma; NA, not available.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download