Abstract
Background
Diacylglycerol O-acyltransferase 2 (DGAT2) synthesizes triacylglycerol (TG) from diacylglycerol; therefore, DGAT2 is considered as a therapeutic target for steatosis. However, the consequence of inhibiting DGAT2 is not fully investigated due to side effects including lethality and lipotoxicity. In this article, we observed the role of DGAT2 in hepatocarcinoma.
Methods
The role of DGAT2 is analyzed via loss-of-function assay. DGAT2 knockdown (KD) and inhibitor treatment on HepG2 cell line was analyzed. Cumulative analysis of cell metabolism with bioinformatic data were assessed, and further compared with different cohorts of liver cancer patients and non-alcoholic fatty liver disease (NAFLD) patients to elucidate how DGAT2 is regulating cancer metabolism.
Results
Mitochondrial function is suppressed in DGAT2 KD HepG2 cell along with the decreased lipid droplets. In the aspect of the cancer, DGAT2 KD upregulates cell proliferation. Analyzing transcriptome of NAFLD and hepatocellular carcinoma (HCC) patients highlights negatively correlating expression patterns of 73 lipid-associated genes including DGAT2. Cancer patients with the lower DGAT2 expression face lower survival rate. DGAT2 KD cell and patients’ transcriptome show downregulation in estrogen- related receptor alpha (ESRRA) via integrated system for motif activity response analysis (ISMARA), with increased dimerization with corepressor prospero homeobox 1 (PROX1).
• DGAT2 inhibition leads to mitochondrial dysfunction in hepatocytes.
• In the progression of NAFLD to HCC, lipid metabolism, including DGAT2, decreases.
• HCC patients with low DGAT2 are linked to mitochondrial dysfunction and low survival rate.
• The ESRRA-PROX1 axis regulates mitochondrial function in hepatocytes.
Since diacylglycerol O-acyltransferases (DGATs) are the final and committed process for triacylglycerol (TG) synthesis, they are concerned as potential therapeutic targets of steatosis [1,2], with a question that which enzyme is contributing more to diseases progression. DGAT2 and DGAT1 share same name but the only equivalence is acyltransferase activity in TG synthesis [3]. Notably, DGAT2 inhibition exhibited significant decreases of lipid accumulation in liver than DGAT1. Therefore, DGAT2 inhibitor is tested as a drug for non-alcoholic fatty liver disease (NAFLD). Multiple studies in rodents illustrates the potential therapeutic action in DGAT2 inhibition for fatty liver; moreover, the phase two clinical trial of DGAT2 inhibitor was performed in 2021 [4]. Thus, DGAT2 is primarily focused as a TG synthesizing enzyme and the other functions are underestimated.
Chronic cirrhosis develops into hepatocellular carcinoma (HCC). While viral hepatitis is well-known etiology, NAFLD is newly emerging etiology for HCC patients [5]. Hepatitis virus B and hepatitis virus C are known as the major cause of liver cancer for accounting about 50% to 70% of HCC patients. However, the development of vaccine and treatments ameliorates the incidence and prevalence of viral infection are keep lowering [6]. Moreover, liver damages with viral hepatitis reverses with sufficient medication. Therefore, viral hepatitis has chances not to progresses into cirrhosis. Unlike viral hepatitis, NAFLD has no specific medication approved by U.S. Food and Drug Administration, and the prevalence of NAFLD increased up to 25% globally in a decade. Consequently, the portion of NAFLD in HCC patients is robustly increased from 5% to maximum 30% in 2000 to 2020s respectively considering cryptogenic cirrhosis in HCC patients [5,7,8]. Due to high prevalence and lack of treatments, NAFLD is more concerned as a key cause of HCC. However, little is known about the role of lipid metabolism in HCC [9-11].
We focused on the correlation of DGAT2 and mitochondria in HCC. Mitochondrial dysfunction is known to be a hallmark of cancer [12-15]. However, the incidence of how mitochondria is facing impairment are not fully understood. With sharing membrane with endoplasmic reticulum, mitochondria are also considered as a hub for lipid synthesis such as phospholipid and even TG despite the well-known functions; a tricarboxylic acid cycle and β-oxidation [16,17]. Thus, proteins located in mitochondrion is highly related to lipid metabolism. We analyzed the impact of DGAT2 knockdown (KD) on hepatoma cell line and evaluated the cell characteristics compared to those of HCC patients. In this article, we have shown the changes of lipid-related genes in NAFLD patients and HCC patients and the impact of DGAT2 KD on mitochondrion genes via downregulation of estrogen-related receptor alpha (ESRRA) activity.
All experiments with cells were processed within 10 passages term. Human hepatoma cell lines, HepG2 and Hep3B, are used in this paper. Media used are Dulbecco’s Modified Eagle Medium (DMEM) with high glucose, glutamine and sodium pyruvate (Thermofisher Scientific, Waltham, MA, USA), supplied with 10% fetal bovine serum (Thermofisher Scientific) and 1% penicillin streptomycin (Thermofisher Scientific). Cell was grown up to 70% to 80% confluency and subcultured into three 100 pi cell culture dish (Sigma-Aldrich, St. Louis, MO, USA). Media was changed for every 3 days.
Basically, overall studies are held with HepG2 cell line. The groups would be HepG2 control shRNA (shCTR) (pLKO.1 shScramble vector) stable cell line and HepG2 DGAT2 shRNA (shDGAT2) (pLKO.1 shDGAT2-TRNC000096) are compared. HepG2 wildtype cell lines are compared with 1 μM DGAT2 inhibitor treated HepG2 cell line. HepG2 shDGAT2 is compared with HepG2 shDGAT2 and ESRRA overexpression model (pCDH-ESRRA overexpression).
Control (SHC002) and shDGAT2 vectors (TRCN0000005195, TRCN0000005196, TRCN0000005197) are purchased from sigma-Aldrich. Lentivirus is produced from HEK293T cell line with transfection of pMDG.2, pRSV, and pMDLg vector with transfer vector. Collected supernatants are filtered with Millex 0.45 μm filter (Sigma-Aldrich). HepG2 cells were subcultured and treated with viral sup and polybrene (Sigma-Aldrich). Transfected cells were selected with puromycin treatments and stored in forzen vials for later use.
Transmission electron microscopy (TEM) is used to detect organelles in cell lines. Target cells are fixed for 24 hours with 2% glutaraldehyde-2% paraformaldehyde in 0.1M phosphate buffer (pH 7.4). Specimen is washed and dehydrated and embedded on Poly/Bed 812 kit (Polysciences, Warrington, PA, USA), and electron microscope oven (TD-700, DOSAKA, Japan) at 65°C for 12 hours. The interest region of block is section in 80 nm and placed on copper grid. The section is then stained with 3% uranyl acetate and 3% lead citrate, final specimen is imaged with a TEM (JEM-1011, JEOL, Tokyo, Japan).
Oxygen consumption rate (OCR) is measured with Agilent XF seahorse mito stress test kit (Agilent Technologies, Santa Clara, CA, USA). The experiments are followed with manufacturer’s protocol. Briefly, 8,000 cells of HepG2 are plated on XFe96 well plate on the day before the assay. Next day, the cells were rinsed with XF DMEM solution (Agilent Technologies) with carbon source added. Drugs are treated with the final concentration of 2.5, 1, and 2.5 μM for oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone/antimycin respectively. For DGAT2 inhibitor treated sample, samples were subcultured with 1 μM of inhibitor. For beta-oxidation test, final concentration of 4 μM etomoxir is treated before oligomycin treatments.
Extracellular acidification rates (ECARs) are calculated via Agilent XF seahorse glycolysis stress test kit (Agilent Technologies). Protocols are performed as manufacturer described. Briefly, HepG2 cell are plated on XFe96 plate. Drugs are treated with 10 mM, 2.5 μM, and 50 mM of glucose, oligomycin and 2-deoxyglucose respectively.
HepG2 cell lines are subcultured on 96 well plate with 5,000 cells/well. Media is changed daily, and cell proliferation rate is measured using cell counting kit-8 (CCK-8) kit (Dojindo, Kumamoto, Japan) following the manufacturer’s protocol. Briefly, 10 μL of CCK-8 solution is added on each plate and incubated on 37°C for 1 hour. The plate is then measured with colorimeter to get absorbance rate.
Raw fastq files are processed quality check and trimming with Trim Galore. The reads are aligned and paired with Bowtie2, samtools, Picard and shell script.MACS2, and Chipseeker packages are used for peak calling and annotation. The following processes are performed via MACROGEN (Seoul, Korea).
Transcriptomic data are collected either from database or lab made libraries. In this article we utilized two RNA sequencing data comparing control HepG2 cell line with HepG2 DGAT2 KD cell line, and control HepG2 cell line vs. HepG2 cell line with DGAT2 inhibitor treated samples. From public database, The Cancer Genome Atlas (TCGA) of liver cancer patients (normal tissue=50, HCC patients=371) data and GSE9632 (NAFLD), GSE135251, GSE1141198, and GSE193084 are analyzed.
HepG2 DGAT2 suppressed models’ RNA is extracted via TRIzol treatment and processed through next generation sequencing. Produced FASTQ files are performed through fastqc and trimmomatic. The reads counts are aligned via HISAT2, Bowtie2, and StingTie. The following processes are performed via MACROGEN. Raw count data are processed to DESeq2 to find differentially expressed genes (DEGs) with false discovery rate (FDR) <0.1, fold change >1.5. Overall gene set enrichments were calculated either Gene Set Enrichment Analysis (GSEA) or gProfiler to find highly significant pathways.
All statistical analyses were performed based on the experiment method and characteristics of data. If the data fits in normal distribution, parametric methods are used and if not, non-parametric methods are used. Normality test is performed with Shapiro’s test. For parametric test, Student’s t-test is used. For nonparametric test, mostly Mann-Whitney U test is utilized for calculating significance of correlation between two factors. Also, spearman correlation, long rank test, one-way analysis of variance (ANOVA) and FDR is used for specific methods and are mentioned in the figure legends. For the methods not indicated in following section are described in Supplementary Methods.
As a family of acyltransferase group, the primary role of DGAT2 is a TG synthesis. We inhibited DGAT2 activity in HepG2 cell line via DGAT2 inhibitor [18], and shDGAT2 (Supplementary Fig. 1A-E). Inhibition of DGAT2 suppresses the production of lipid droplets while 200 mM of oleic acid and palmitic acid are treated (Fig. 1A and B). Along the decrease of lipid droplet, depletion of DGAT2 enzyme markedly diminished mitochondria (Fig. 1C). The relative mitochondria area in cytosol, and the distribution of mitochondrial length suggests significant decrease in its size (Fig. 1D and E). This change is not associated with only morphologies, but the mitochondrial DNA contents and membrane potentials are also decreased (Fig. 1F and G). We assumed the dysregulation in mitochondrial dynamics mediated the morphologies, and were able to detect upregulated fission proteins, fission, mitochondrial 1 (FIS1), and phosphorylation status of Ser616 of dynamin-related protein 1 (pDRP1; Ser616), in DGAT2 KD HepG2 cell line (Fig. 1H) [19]. Consequently, the OCR, especially the maximal respiration rate, is suppressed (Fig. 1I). Moreover, KD of DGAT2 suppressed β-oxidation that etomoxir, carnitine palmitoyl transferase 1 inhibitor, treatments resulted no difference in OCR while control HepG2 resulted significant decrease in OCR (Fig. 1J). Altogether, suppression of DGAT2 activity in HepG2 cell line promotes mitochondrial dysfunction along the decrease of lipid accumulation and eventual decrease of cellular adenosine triphosphate (ATP) level (Fig. 1K).
Lipid accumulation in liver upon 5% of total weight are considered as liver steatosis. However, the lipid accumulation is known to decrease in cirrhotic phase. However, the consequences of lipid metabolism in HCC patients are not fully understood. Thus, we analyzed two datasets, NAFLD patients (GSE89632), and HCC patients from TCGA database to find out DEGs in between two cohorts (Fig. 2A). Among four groups of DEGs (Supplementary Fig. 2), 805 DEGs were upregulated in NAFLD patients while downregulated in HCC patients, is the only group to exhibit lipid-related genes (Fig. 2B). This clues that even HCC exhibits suppression in lipid metabolism. Thus, we categorized the patients into different group via spearman correlation based on 73 lipid genes. The heatmap categorized HCC patients into three groups. Group 1 are showing highest correlation with adjacent normal tissue, where group 3 exhibits the lowest correlation with the adjacent tissue (Fig. 2C). The clear separation of group 1 and group 3 in principle component analysis plot suggests metabolic differences in between two, while group 3 is shown to has lower survival rate than group 1 (Fig. 2D and E). Notably, the expression level of DGAT2 is highly correlating with the relationship of NAFLD and HCC patients and gradually decreases with the severity (Fig. 2F and G). The spatial data [20,21] exhibits the DGAT2 expression patterns in liver that the level of DGAT2 is upregulated in the region of ectopic fat in steatosis patients, marked with black arrows (Fig. 2G). Moreover, similar outcome is detected from different cohorts that the expression level of DGAT2 decreases as patients progress into cirrhosis and HCC (Fig. 2H). Next, we have performed bulk RNA sequencing for two DGAT2 inhibition models and were able to find that the correlations of downregulated DEGs in our model and HCC patients come from the gene set; GO:CC mitochondrion (Fig. 2I). Therefore, these data draw the correlation between HCC severity with lipid metabolism and mitochondrial functions, especially with DGAT2.
To elucidate the activity of DGAT2, HCC patients are grouped into high DGAT2 expressing group and low DGAT2 expressing group (Fig. 3A and B). Considering that DGAT2 gradually decreases as HCC progresses based on modified tumor-node-metastasis staging (Fig. 3C), patients with low DGAT2 expression tends to show lower survival rate (Fig. 3D). Analysis of DEGs suggest the alteration in the genes regulating mitochondrial dynamics. In low DGAT2 group, fusion inhibiting vesicle amine transporter 1 and dynamin 1 like are upregulated while mitochondrial biogenesis related signal transducer and activator of transcription 2 are downregulated. These imply modification of mitochondrial dynamics (Fig. 3E). Since mitochondrial activity negatively correlates with glycolysis, we run GSEA for probable metabolic changes. The results highlighted upregulation of glycolysis and cell cycle, and downregulation of fatty acid metabolism (Fig. 3F). Therefore, HCC patients exhibited the correlation of DGAT2 and mitochondrial dysfunction.
In terms of cancer profile, HepG2 exhibits enhanced cell proliferation rate while DGAT2 activity is suppresses via inhibitor or shRNA (Fig. 4A). Cell cycle analysis with flow cytometry and propidium iodide shows increased S and G2 phase in KD cell line (Fig. 4B). Moreover, protein level of cyclin-dependent kinase 2, cyclin A2, and cyclin E1 are upregulated in DGAT2 KD model (Fig. 4C). Due to upregulated cell proliferation, DGAT2 KD model showed higher speed in wound closure (Fig. 4D and E). Moreover, not only in HepG2 cell line but Hep3B cell line, which expresses low level of DGAT2, exhibits significantly higher cell proliferation rate at day 2 (Fig. 4F and G). This evidence highlights that the DGAT2 mediated mitochondrial dysfunction accelerates cell proliferation, and it can be generalized in other HCC cell lines.
These correlation of DGAT2 expression level and cell proliferation are predicted to be the consequence of upregulated glycolysis in DGAT2 inhibited condition. When DGAT2 activity is suppresses either via inhibitor or shRNA, the final product of glycolysis, lactate, level is increased by 150% of control (Fig. 4H). In addition, ECAR showed the level of glycolysis is increased when DGAT2 is inhibited with 1 μM of PF-06424439 (Fig. 4I and J). As predicted, only DGAT2 KD HepG2 showed suppression in cell growth rate, while it was tolerable for control group. Thus, mitochondrial dysfunction by DGAT2 suppression shifts metabolism toward glycolysis and induces cell proliferation in HepG2 [22].
To distinguish a key regulator of metabolic changes, motif activities were calculated with web-based tool, integrated system for motif activity response analysis (ISMARA). The prediction brought three motifs out of top 10 motifs that activated in NAFLD patients but deactivated in HCC patients. Among three motifs, PLAG1 like zinc finger 1, ESRRA, and KLF transcription factor 16, ESRRA fits for the regulating mitochondrial homeostasis and biogenesis (Fig. 5A) [23,24]. ESRRA motif activity from transcriptome of both DGAT2 inhibitor and shDGAT2 model were calculated to be suppresses (Fig. 5B), which confirmed with the luciferase assay we performed. To find motif activity and downstream gene translation. With the insertion of p3x estrogen-related response element (p3xERRE) vector tagged with firefly luciferase, DGAT2 KD model showed half of a relative luciferase activity of control cell (Fig. 5C). However, the only known endogenous ligand of ESRRA, the cholesterol level showed no differences, implying other factors are contributing on the activity of ESRRA (Fig. 5D) [25]. Yet, transcriptome data suggest overall downregulation on the genes with estrogen-related receptor response element (ERRE) sequences and the peak targeted on ERRE are shown to be decreased from analysis of assay for transposase-accessible chromatin sequencing of shDGAT2 model (Fig. 5E and F). Thus, we conducted subcellular fraction to divide nucleus from cytosol. According to Western blot, the amount of ESRRA translocated in nucleus are diminished (Fig. 5G). Notably, immunoprecipitation via ESRRA-antibody from nucleus fraction suggested decreased ESRRA and increased prospero homeobox 1 (PROX1) bound to ESRRA (Fig. 5H). PROX1 is a suppressor of ESRRA; therefore, the transcriptional network of ESRRA-PROX1 deactivates the ESRRA and inhibits downstream gene transcription [26].
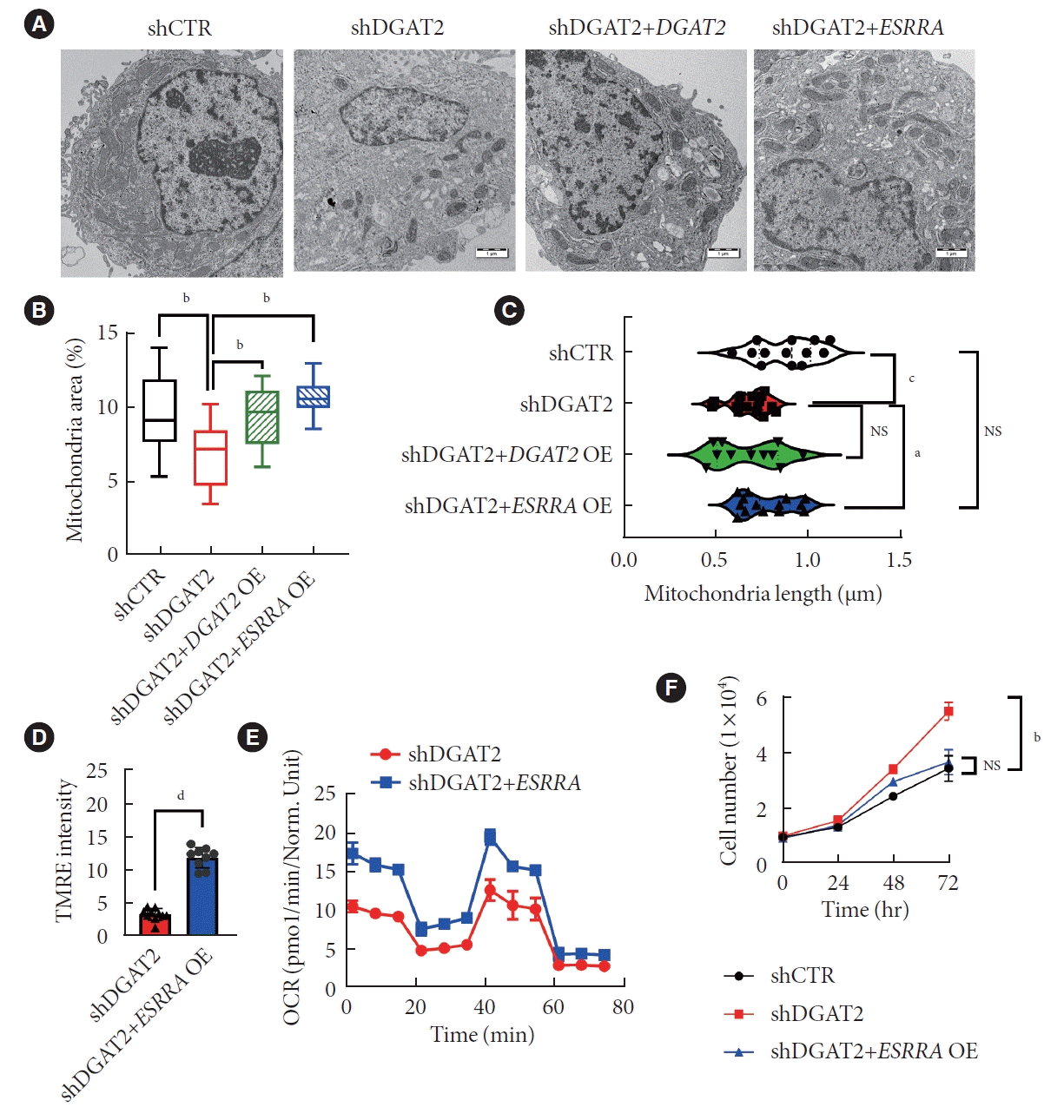
To find out whether downregulated ESRRA is promoting mitochondrial dysfunction, DGAT2 KD HepG2 model is overexpressed with the ESRRA construct. Two days after transfection, cells were performed TEM to assess mitochondrial morphologies (Fig. 6A). Mitochondrial area and length are decreased upon DGAT2 KD and is recued via ESRRA overexpression (Fig. 6B and C). As morphology is rescued, mitochondrial potential is also rescued (Fig. 6D). Following with the increase of OCR via ESRRA overexpression, mitochondrial dysfunction is ameliorated via ESRRA overexpression (Fig. 6E). Moreover, the cell proliferation rate decreased toward control cell compared to the DGAT2 KD model (Fig. 6F). Thus, upregulation of suppressed ESRRA activity reversed mitochondrial function and cell proliferation of HepG2 cell.
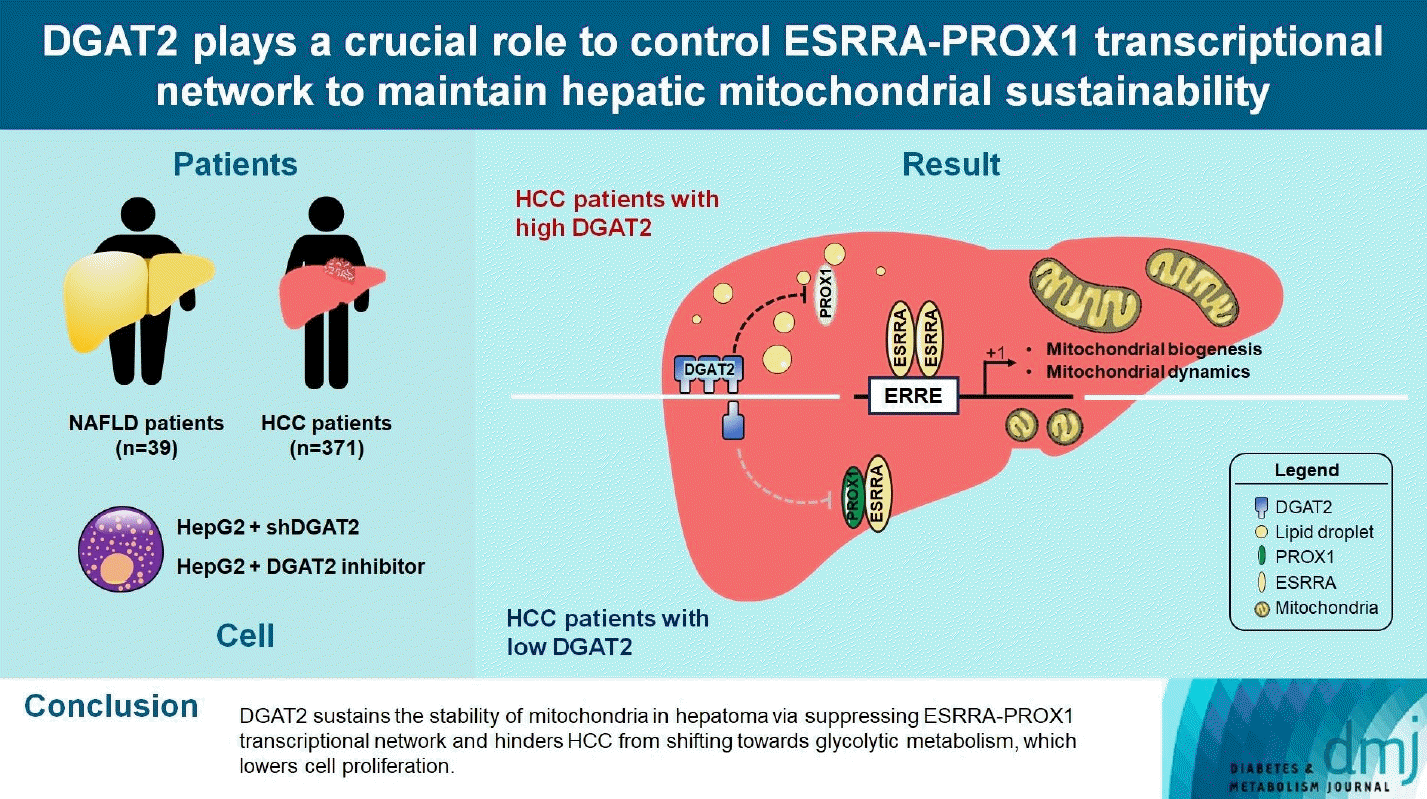
We have presented inhibition of DGAT2, TG synthesizing enzyme, downregulates mitochondrial function via transcriptional network of ESRRA-PROX1. Mitochondria is diminished and cell proliferation is upregulated, which leads to the severity of HCC. Current understanding of mitochondria exceeds its classical role as a cellular powerplant, and further researched as an organelle regulating reactive oxygen species, building blocks, and hypoxia related signals [12]. Although we mainly focused on degradation of mitochondria and classical Warburg effects, upregulated fission could be considered as an advantageous feature for the cancer survival [27]. Fusion is a process of mitochondria to mix damaged mitochondria with healthy one for diluting the damages. Upregulated fission over fusion causes increased stress and promotes mitophagy, which produces energy and biomolecules for cell growths [28]. In TEM of DGAT2 KD cell, upregulated mitophagy is detected, which implies the possible sources for cell proliferation and autophagosome markers are elevated (Supplementary Fig. 3).
While DGAT2 links NAFLD and HCC, it is hard to specify the consequences of a NAFLD on HCC since each patient possesses multiple etiologies. Based on TCGA database of Liver Hepatocellular Carcinoma (LIHC) patients, the number of patients with single etiology are seven for NAFLD. Moreover, usually patients are categorized as non B viral and non C viral (NBNC), which is not actually a NAFLD patients but also includes type 2 diabetic patients and alcoholic fatty liver diseases. It could be the reason but the DGAT2 expression level is not distinguishable according to the etiologies (Supplementary Fig. 4A). The correlation of DGAT2 and other clinical factors from TCGA-LIHC sample suggests higher DGAT2 expression in female; however, no correlation is detected with body mass index (Supplementary Fig. 4B and C). Several studies suggest the correlation of liver fibrosis above F3 are highly correlated with the prevalence of HCC [29]. Notable point is that expression level of DGAT2 not only correlates with NAFLD and HCC, but also negatively correlates with fibrosis level (Supplementary Fig. 4D and E). Therefore, further studies are required to determine when, and which factors regulate the expression of DGAT2 in liver disease [30].
One clue from TCGA data exhibits possible factor for DGAT2 downregulation. Analyzing 50 paired HCCs with corresponding adjacent tissue points that average DGAT2 expression level of adjacent tissues does not represent those of paired cancer; rather, the expression level seems to change during tumor formation (Supplementary Fig. 5A). To figure out possible factors regulating DGAT2 expression, methylation, copy number variant and mutation were evaluated. While copy number variants have no differences, methylation on CpG island of DGAT2 were significantly higher in low DGAT2 expressing group (Supplementary Fig. 5B and C). For mutation, although DGAT2 mutation is known to promote Charcot-Marie-Tooth disease [31], DGAT2 is silently mutated in only two patients out of 371 patients (Supplementary Fig. 5D). It can be implied that HCC obtain methylation during tumor formation, which determines the expression level of DGAT2.
In this paper, we proposed the downregulation of ESRRA as a driving force for mitochondrial dysfunction; however, the direct connection of DGAT2 and ESRRA is still in vague. Probable change from DGAT2 inhibition is the metabolite changes, especially diacylglycerol (DAG). The assay of DGAT2 KD cell line suggest no significant increase in DAG concentration, rather exhibits slight decreased concentration, correlates with increased concentration of phosphatidic acid (PA) level (Supplementary Fig. 1F and G) [32]. DAG is converted into PA via diacylglycerol kinase and used as a derivative of plasma membrane. On the other hand, protein kinase C (PRKC) family has C1 domains for DAG binding for its activation [33-35]. Among PRKC family, PRKC epsilon is downregulated, and it is known to activate ESRRA via phosphorylation on T106, S110, and T124 [36]. Likewise, the cofactor of ESRRA needs further evaluation. PPARGC1A (PGC1A) is one well-known coactivator of ESRRA, while PROX1 is a corepressor of ESRRA. The protein level of PGC1A is significantly downregulated upon DGAT2 KD. In contrast the expression of PROX1 is upregulated (Supplementary Fig. 6A). Moreover, the KD of PROX1 suppressed cell proliferation of DGAT2 KD cell (Supplementary Fig. 6B).
Immune response in tumor microenvironments regulates the growth and survival rate of tumor and immune evasion is considered as a key factor in cancer hallmarks. Little is known about the correlations between DGAT2 and immune function in cancer. Moreover, the controversy perspectives of immune response in NAFLD and HCC make it harder to comprehend that in NAFLD, anti-immune functions are considered as a positive marker where in HCC, pro-immune functions are considered as safer. Based on immune signature, pro- and anti-immune activities are scored [37]. The enrichment score suggests both pro-inflammatory signature, naïve and cytotoxic signatures, and antiinflammatory signature, exhaustion and regulatory T-cell signatures, are upregulated in low DGAT2 group (Supplementary Fig. 7A). Therefore, considering single factor make it hard to interpret the correlation of DGAT2 and immune function. However, comparing the cytotoxic signature with exhaustion signature, only low DGAT2 group exhibits significantly increased exhaustion signature than cytotoxic signature (Supplementary Fig. 7B).
Metabolism shift in the cancer is an essential adaptation for cancer survival. However, the role of lipid in HCC is yet to be clarified due to diversity of lipid categories. We revised HCC in terms of lipogenesis and how mitochondrial function be modulated via DGAT2 and suppresses cancer cell proliferation.
SUPPLEMENTARY MATERIALS
Supplementary materials related to this article can be found online at https://doi.org/10.4093/dmj.2023.0368.
Supplementary Fig. 1.
Construction of diacylglycerol O-acyltransferase 2 (DGAT2) suppression in vitro model. (A) Brief scheme for DGAT2 suppression model via inhibitor and shRNA. (B) Relative DGAT2 expression after shRNA treatments. (C) DGAT2 protein expression in TRCN0000005196 transfection. Asterisk is to indicate non-specific band. (D) BODIPY 493/503 stained lipid droplet after DGAT2 inhibitor PF-06424439 (Pfizer) treatment. (E) Relative intensity of BODIPY after inhibitor treatments. (F) Cellular diacylglycerol (DAG) concentration after DGAT2 suppression. (G) Cellular phosphatidic acid level after DGAT2 suppression. Data represent mean±standard deviation. NC, negative control; NS, not significant; shCTR, control shRNA; shDGAT2, DGAT2 shRNA; DAPI, 4΄,6-diamidino-2-phenylindole. aP<0.04, bP<0.01, cP<0.001, dP<0.0001; two-tailed Student΄s t-test.
Supplementary Fig. 2.
Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of differentially expressed genes (DEGs) of non-alcoholic fatty liver disease (NAFLD) patients and hepatocellular carcinoma (HCC) patients. (A) Significantly enriched KEGG pathways of DEGs both upregulated in NAFLD and HCC patients. (B) DEGs both downregulated in NAFLD and HCC patients. (C) DEGs upregulated in NAFLD patients but downregulated in HCC patients. (D) DEGs downregulated in NAFLD patients but upregulated in HCC patients. The significance of following functional profiling is determined via false discovery rate. TNF, tumor necrosis factor; AGE-RAGE, advanced glycation endproducts-receptor for advanced glycation end products; MAPK, mitogen-activated protein kinase; IL, interleukin; JAK-STAT, Janus kinase/signal transducers and activators of transcription; NF, nuclear factor.
Supplementary Fig. 3.
Upregulated autophagosome and mitophagy in HepG2 diacylglycerol O-acyltransferase 2 (DGAT2) knockdown (KD) cell line. (A) Mitophagy is detected in transmission electron microscopy images, and red arrowheads indicate mitochondria in HepG2 cell. Mitophagy is detected only in DGAT2 KD cell line. (B) Western blot image of autophagosome marker lysosomal associated membrane protein 1 (LAMP1) and microtubule associated protein 1 light chain 3 beta (LC3B) with the endoplasmic reticulum marker, calnexin (CANX) as a reference. shCTR, control shRNA; shDGAT2, DGAT2 shRNA.
Supplementary Fig. 4.
Correlation of diacylglycerol O-acyltransferase 2 (DGAT2) and clinical factors. (A) Expression level of DGAT2 based on etiologies. (B) DGAT2 expression level based on the gender in hepatocellular carcinoma patients. (C) Correlation of DGAT2 and body mass index (BMI) from The Cancer Genome Atlas (TCGA)-Liver Hepatocellular Carcinoma (LIHC) cohorts. Calculated with linear regression. (D) Expression level of DGAT2 based on liver pathology state in GSE193084. (E) Expression level of DGAT2 based on fibrosis score of patients in GSE193084. Data represent mean±standard deviation. NAFLD, non-alcoholic fatty liver disease; NS, not significant. aP<0.05, bP<0.01, cP<0.0001; two-tailed Mann-Whitney U test.
Supplementary Fig. 5.
Genomic and epigenomic data of hepatocellular carcinoma (HCC) patients. (A) Diacylglycerol O-acyltransferase 2 (DGAT2) expression level of patients paired with adjacent normal tissue. (B) Methylation level of CpG island on DGAT2. (C) Copy number variant of DGAT2 in HCC patients. (D) Number of genes according to the number of patients with mutation. DGAT2 is silently mutated in two patients. Data represent mean±standard deviation. NS, not significant; SNP, single nucleotide polymorphism; TCGA, The Cancer Genome Atlas. aP<0.01, bP<0.0001: two-tailed Student’s t-test in (B, C); two-tailed Mann-Whitney U test in (A).
Supplementary Fig. 6.
Expression level of estrogen-related receptor alpha (ESRRA) cofactors correlate with the downregulated transcriptional activity of ESRRA. (A) Protein level of ESRRA and cofactors (PGC1A and prospero homeobox 1 [PROX1]) are assessed, which the coactivator PGC1A level is decreased and corepressor PROX1 level is up regulated. Asterisk indicates nonspecific bands (110 kDa). (B) Cell proliferation level of HepG2 diacylglycerol O-acyltransferase 2 (DGAT2) knockdown cell line suppresses after PROX1 inhibition. Data represent mean±standard deviation. shCTR, control shRNA; shDGAT2, DGAT2 shRNA. aP<0.001, bP<0.0001 two-tailed Student’s t-test.
Supplementary Fig. 7.
Diacylglycerol O-acyltransferase 2 (DGAT2) xpression and immune characteristics of hepatocellular carcinoma patients. (A) DGAT2 high group and low group are compared for their immune signatures, including cytotoxic signature, exhaustion signature, naïve signature, regulatory T-cell signature, monocyte signature, M1 signature, and M2 signature. (B) Comparison of cytotoxic signature with exhaustion signature infers higher exhaustion signature in low DGAT2 group. Data represent mean±standard deviation. aP<0.05, bP<0.01, cP<0.0001 two-tailed Mann-Whitney U test.
Notes
AUTHOR CONTRIBUTIONS
Conception or design: Y.L., S.F., J.W.K.
Acquisition, analysis, or interpretation of data: all authors.
Drafting the work or revising: Y.L., S.F.
Final approval of the manuscript: S.F., J.W.K.
FUNDING
This research is funded by National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (NRF-2018R1A5A2025079); and supported by grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health &Welfare, Republic of Korea (grant number: HI14 C1324).
ACKNOWLEDGMENTS
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas (TCGA) Research Network: https://www.cancer.gov/tcga. This study was carried out in part in the Yonsei Advanced Imaging Center in cooperation with Carl Zeiss Microscopy, Yonsei University College of Medicine.
REFERENCES
1. Jin Y, McFie PJ, Banman SL, Brandt C, Stone SJ. Diacylglycerol acyltransferase-2 (DGAT2) and monoacylglycerol acyltransferase-2 (MGAT2) interact to promote triacylglycerol synthesis. J Biol Chem. 2014; 289:28237–48.
2. Yenilmez B, Wetoska N, Kelly M, Echeverria D, Min K, Lifshitz L, et al. An RNAi therapeutic targeting hepatic DGAT2 in a genetically obese mouse model of nonalcoholic steatohepatitis. Mol Ther. 2022; 30:1329–42.

3. Chitraju C, Walther TC, Farese RV Jr. The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J Lipid Res. 2019; 60:1112–20.

4. Calle RA, Amin NB, Carvajal-Gonzalez S, Ross TT, Bergman A, Aggarwal S, et al. ACC inhibitor alone or co-administered with a DGAT2 inhibitor in patients with non-alcoholic fatty liver disease: two parallel, placebo-controlled, randomized phase 2a trials. Nat Med. 2021; 27:1836–48.

5. Singal AG, El-Serag HB. Rational HCC screening approaches for patients with NAFLD. J Hepatol. 2022; 76:195–201.

6. Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022; 77:1598–606.

7. Nagaoki Y, Hyogo H, Ando Y, Kosaka Y, Uchikawa S, Nishida Y, et al. Increasing incidence of non-HBV- and non-HCV-related hepatocellular carcinoma: single-institution 20-year study. BMC Gastroenterol. 2021; 21:306.

8. Song K, Yang J, Lee HS, Kim SJ, Lee M, Suh J, et al. Changes in the prevalences of obesity, abdominal obesity, and non-alcoholic fatty liver disease among Korean children during the COVID-19 outbreak. Yonsei Med J. 2023; 64:269–77.

9. Loomba R, Lim JK, Patton H, El-Serag HB. AGA clinical practice update on screening and surveillance for hepatocellular carcinoma in patients with nonalcoholic fatty liver disease: expert review. Gastroenterology. 2020; 158:1822–30.

10. Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017; 16:76.

11. Bae H, Lee SA, Choi JW, Hwang SH, Park S, Park MS. Effectiveness of hepatocellular carcinoma surveillance and an optimal surveillance interval: nationwide cohort of Korea. Yonsei Med J. 2021; 62:758–66.

12. Lee YG, Park DH, Chae YC. Role of mitochondrial stress response in cancer progression. Cells. 2022; 11:771.

13. Grasso D, Zampieri LX, Capeloa T, Van de Velde JA, Sonveaux P. Mitochondria in cancer. Cell Stress. 2020; 4:114–46.

14. Yambire KF, Fernandez-Mosquera L, Steinfeld R, Muhle C, Ikonen E, Milosevic I, et al. Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases. Elife. 2019; 8:e39598.

16. Eynaudi A, Diaz-Castro F, Borquez JC, Bravo-Sagua R, Parra V, Troncoso R. Differential effects of oleic and palmitic acids on lipid droplet-mitochondria interaction in the hepatic cell line HepG2. Front Nutr. 2021; 8:775382.

17. Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015; 13:89.

18. Futatsugi K, Kung DW, Orr ST, Cabral S, Hepworth D, Aspnes G, et al. Discovery and optimization of imidazopyridine-based inhibitors of diacylglycerol acyltransferase 2 (DGAT2). J Med Chem. 2015; 58:7173–85.

19. Xie L, Shi F, Li Y, Li W, Yu X, Zhao L, et al. Drp1-dependent remodeling of mitochondrial morphology triggered by EBVLMP1 increases cisplatin resistance. Signal Transduct Target Ther. 2020; 5:56.

20. Wu R, Guo W, Qiu X, Wang S, Sui C, Lian Q, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021; 7:eabg3750.

21. Habenicht LK, Wang Z, Zhang X, Li Y, Mogler C, Huspenina JS, et al. The C1q-ApoE complex: a new hallmark pathology of viral hepatitis and nonalcoholic fatty liver disease. Front Immunol. 2022; 13:970938.

22. Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006; 25:4633–46.

23. Fox SN, McMeekin LJ, Savage CH, Joyce KL, Boas SM, Simmons MS, et al. Estrogen-related receptor gamma regulates mitochondrial and synaptic genes and modulates vulnerability to synucleinopathy. NPJ Parkinsons Dis. 2022; 8:106.

24. Hong EJ, Levasseur MP, Dufour CR, Perry MC, Giguere V. Loss of estrogen-related receptor α promotes hepatocarcinogenesis development via metabolic and inflammatory disturbances. Proc Natl Acad Sci USA. 2013; 110:17975–80.

25. Yi SW, Kim SH, Han KJ, Yi JJ, Ohrr H. Higher cholesterol levels, not statin use, are associated with a lower risk of hepatocellular carcinoma. Br J Cancer. 2020; 122:630–3.

26. Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, et al. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 2010; 24:537–42.
27. Tang M, Yang M, Wu G, Mo S, Wu X, Zhang S, et al. Epigenetic induction of mitochondrial fission is required for maintenance of liver cancer-initiating cells. Cancer Res. 2021; 81:3835–48.

28. Lu M, van Tartwijk FW, Lin JQ, Nijenhuis W, Parutto P, Fantham M, et al. The structure and global distribution of the endoplasmic reticulum network are actively regulated by lysosomes. Sci Adv. 2020; 6:eabc7209.

29. Sunami Y. NASH, fibrosis and hepatocellular carcinoma: lipid synthesis and glutamine/acetate signaling. Int J Mol Sci. 2020; 21:6799.

30. Gluchowski NL, Gabriel KR, Chitraju C, Bronson RT, Mejhert N, Boland S, et al. Hepatocyte deletion of triglyceride-synthesis enzyme acyl CoA: diacylglycerol acyltransferase 2 reduces steatosis without increasing inflammation or fibrosis in mice. Hepatology. 2019; 70:1972–85.

31. Hong YB, Kang J, Kim JH, Lee J, Kwak G, Hyun YS, et al. DGAT2 mutation in a family with autosomal-dominant earlyonset axonal charcot-marie-tooth disease. Hum Mutat. 2016; 37:473–80.
32. Cai J, Abramovici H, Gee SH, Topham MK. Diacylglycerol kinases as sources of phosphatidic acid. Biochim Biophys Acta. 2009; 1791:942–8.

33. Yang C, Kazanietz MG. Divergence and complexities in DAG signaling: looking beyond PKC. Trends Pharmacol Sci. 2003; 24:602–8.

34. Kolczynska K, Loza-Valdes A, Hawro I, Sumara G. Diacylglycerol-evoked activation of PKC and PKD isoforms in regulation of glucose and lipid metabolism: a review. Lipids Health Dis. 2020; 19:113.

35. Cooke M, Magimaidas A, Casado-Medrano V, Kazanietz MG. Protein kinase C in cancer: the top five unanswered questions. Mol Carcinog. 2017; 56:1531–42.

Fig. 1.
Downregulation of diacylglycerol O-acyltransferase 2 (DGAT2) in HepG2 mitigates mitochondrial function. (A) Oleic acid and palmitic acids are treated for 200 μM each, 4 hours incubated in HepG2 models. (B) Intensity of BODIPY 493/503 staining of lipid droplets, normalized with 4΄,6-diamidino-2-phenylindole (DAPI), exhibits suppressed lipid synthesis via DGAT2 inhibition. (C) Mitochondrial transmission electron microscopy photo (×10,000) and mitotracker staining for mitochondrial morphology detection. (D) Quantified mitochondrial area based on a cytosol area. (E) Distribution of mitochondrial length across the cytosol. (F) Tetramethylrhodamine, ethyl ester (TMRE) intensity indicates the membrane potential of mitochondria. (G) Mitochondrial DNA relative to the reference DNA indicates decreased in DGAT2 knockdown HepG2. (H) Western blot data of fission- related proteins. (I) Oxygen consumption rate (OCR) rate of HepG2 cell line after DGAT2 suppression. (J) OCR changes after carnitine palmitoyl transferase I (CPT1) inhibition via etomoxir. (K) Cellular adenosine triphosphate level. Data represent mean±standard deviation. OAPA, oleic acid and palimatic acid; shDGAT2, DGAT2 shRNA; shCTR, control shRNA; FIS1, fission, mitochondrial 1; pDRP S637, phosphorylayion status of Ser-637 of dynamin-related protein 1; MFN1, mitofusin 1; NS, not significant. aP<0.05, bP<0.01, cP<0.001, dP<0.0001; two-tailed Student’s t-test.
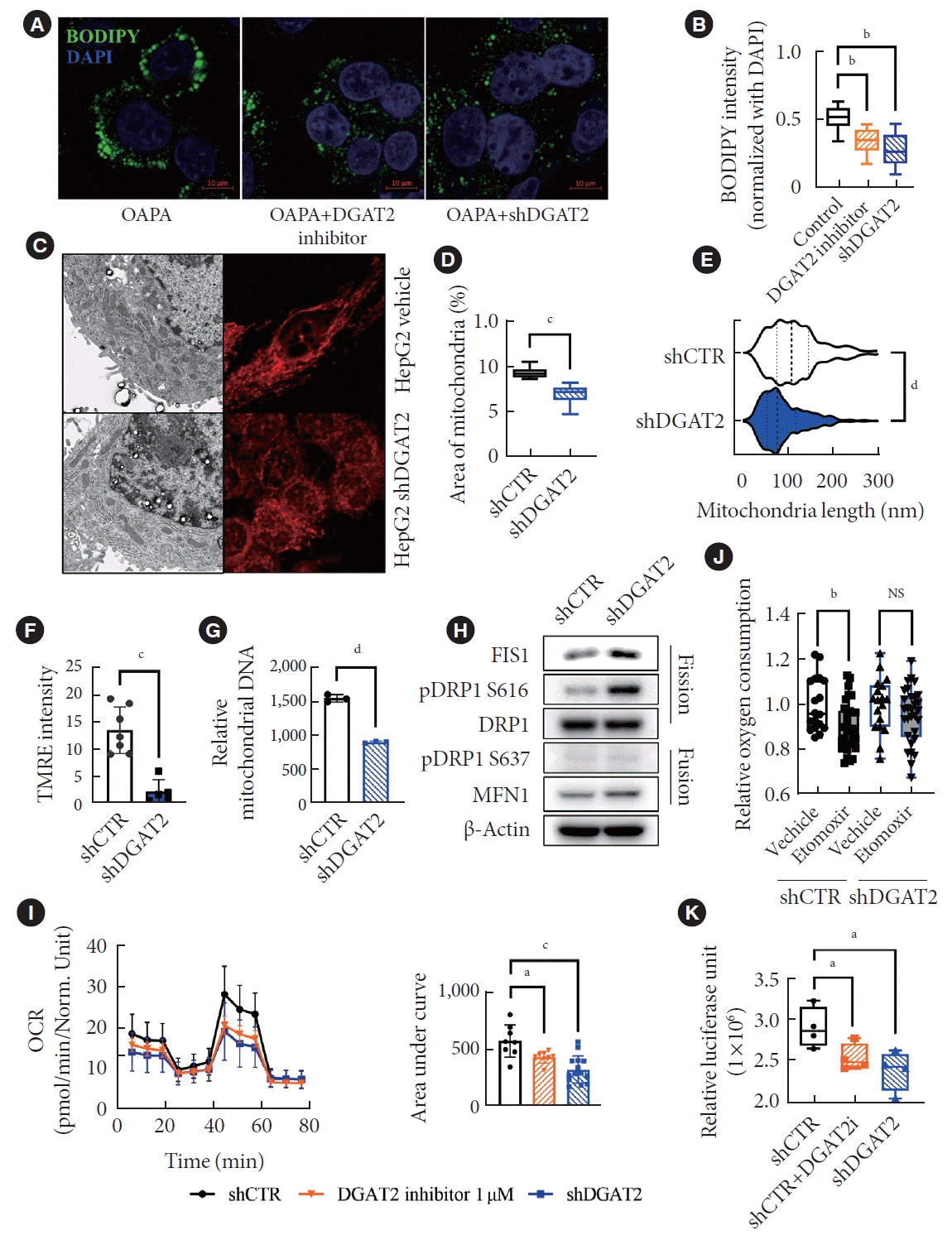
Fig. 2.
Lipid accumulation drops dramatically in hepatocellular carcinoma (HCC) compares to non-alcoholic fatty liver disease (NAFLD) including downregulation of diacylglycerol O-acyltransferase 2 (DGAT2). (A) Vann diagram of differentially expressed genes (DEGs) of NAFLD and HCC patients compare to the normal. Highlights four groups of DEGs. (B) Enrichment scores based on Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set were calculated based on false discovery rate. Lipid-related pathways were only present in the group 0f 805 genes, which upregulated in NAFLD patients and downregulated in HCC patients. (C) Lipid-related genes categorizes HCC patients in to three groups according to the spearman correlation. (D) Principal component analysis distinctly divides group 1 and group 3. (E) Overall survival rate is drawn via Kaplan-Meier plot. Group 3 exhibits lower survival rate than that of group 1. (F) Normalized expression level of DGAT2 is assessed from spatial data of steatosis and HCC patients, which increases in steatosis but decreases in HCC patients. (G) Spatial data compares expression of DGAT2 to liver histology images. Arrows indicate that DGAT2 expression is increased in ectopic fat regions. (H) Expression level of DGAT2 increases in NAFLD while decreases in HCC, and further decreases in group 3. (I) Downregulated DEGs from DGAT2 suppression models correlates with the downregulated pathways in HCC patients only when DEGs are related with mitochondria. Data represent mean±standard deviation. P value for Kaplan-Meier plot is calculated via log rank test. TCGA, The Cancer Genome Atlas; LIHC, Liver Hepatocellular Carcinoma; FA, fatty acid; PC, principle component; NS, not significant; shDGAT2, DGAT2 shRNA. aP<0.05, bP<0.001, cP<0.0001; two-tailed Mann-Whitney U test.
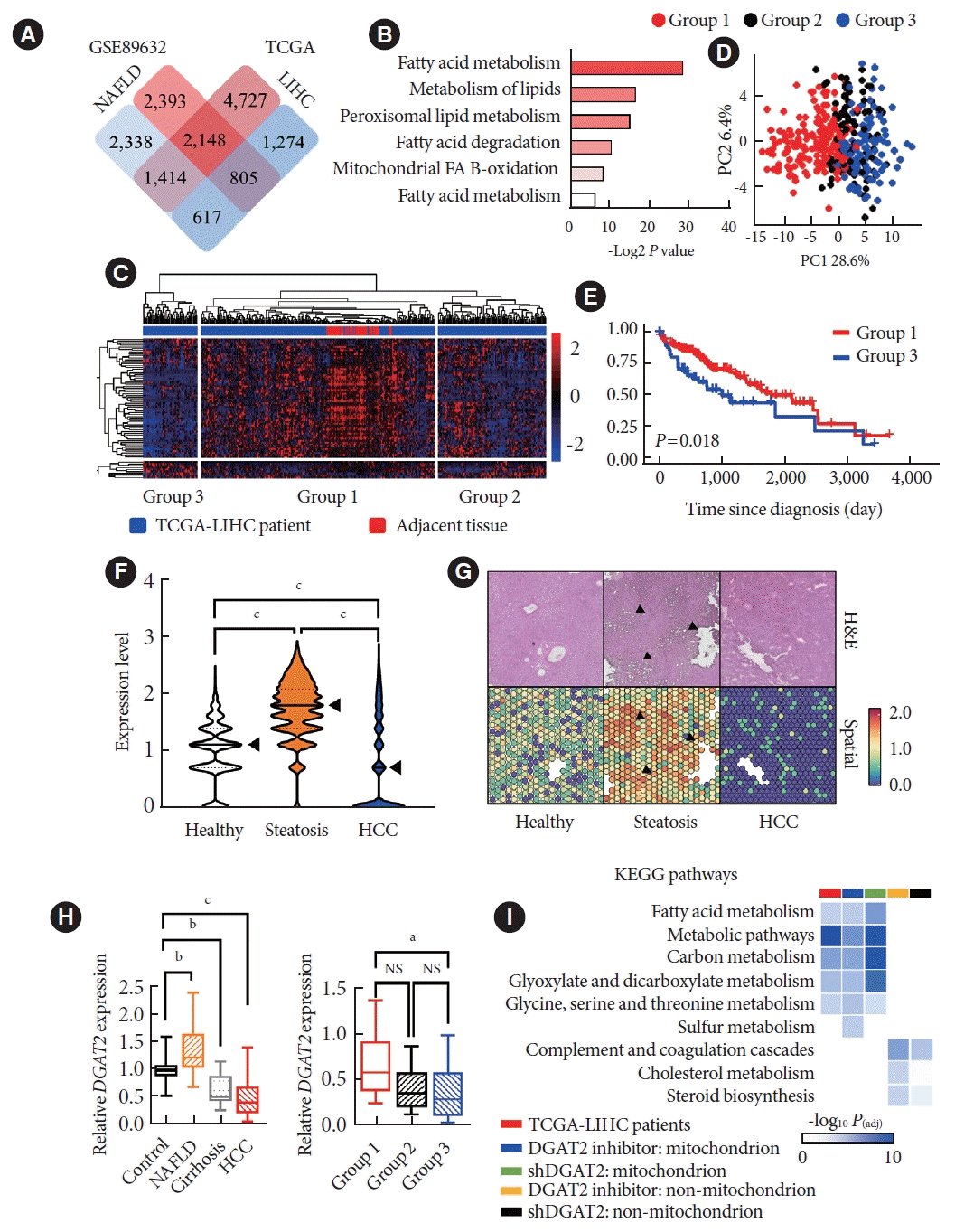
Fig. 3.
Diacylglycerol O-acyltransferase 2 (DGAT2) low expressing patients has lower survival rates with mitochondrial dysfunctions. (A) DGAT2 expression level of high DGAT2 expressing group (n=100) and low DGAT2 expressing group (n=100). (B) DGAT2 expression level according to the stage of hepatocellular carcinoma (HCC) patients with non-parametric one-way analysis of variance (ANOVA) test. (C) Principal component analysis of high and low DGAT2 expressing group. (D) Overall survival rate of high and low DGAT2 expressing group. (E) Heatmap of mitochondrial dynamic related genes. (F) Gene set enrichment of HCC patients, high DGAT2 expressing patients versus low DGAT2 expressing patients. Data represent mean±standard deviation. P value for Kaplan-Meier plot is calculated via log rank test. Significance of Gene Set Enrichment Analysis (GSEA) data is analyzed with false discovery rate calculations. PC, principle component; VAT1, vesicle amine transporter 1; COX10, cytochrome c oxidase assembly factor heme A:farnesyltransferase COX10; DNM1L, dynamin 1 like; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1; STAT2, signal transducer and activator of transcription 2; KEGG, Kyoto Encyclopedia of Genes and Genomes. aP<0.0001; two-tailed Mann-Whitney U test.
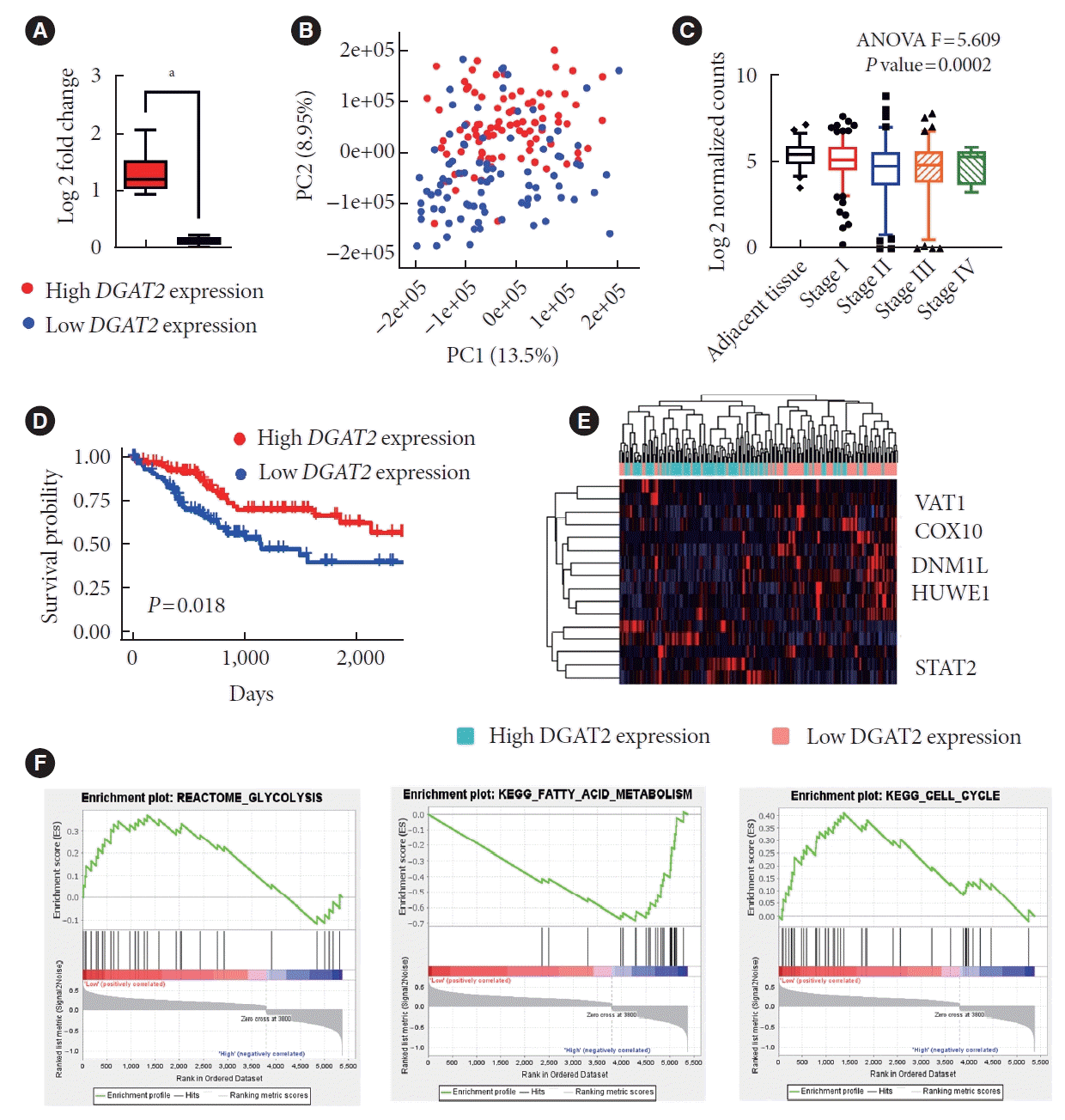
Fig. 4.
Mitochondrial dysfunction and upregulated glycolysis promote cell proliferation. (A) HepG2 cell proliferation after diacylglycerol O-acyltransferase 2 (DGAT2) suppression. (B) Cell cycle of DGAT2 knockdown (KD) cell. (C) Western blot data of cell cycle regulating proteins. (D) Graph of HepG2 wound closure shows migration ability of DGAT2 KD HepG2 cell line. (E) Microscopic image of HepG2 wound healing assay. (F) Cellular lactate level after DGAT2 inhibition. (G) Extracellular acidification rate (ECAR) after DGAT2 inhibitor treatments. (H) DGAT2 expression level of different hepatocellular carcinoma cell lines; HepG3; Hep3B. (I) Cell proliferation rate of Hep3B and HepG2. (J) Glycolysis inhibition inhibits cell proliferation of DGAT2 KD HepG2 cell only. Data represent mean±standard deviation. shCTR, control shRNA; shDGAT2, DGAT2 shRNA; PI_A, propidium iodide_area; CDK2, cyclin-dependent kinase 2; CCNA2, cyclin A2; CCNB1, cyclin B1; CCNE1, cyclin E1; 2-DG, 2-deoxy-D-glucose. aP<0.05, bP<0.01, cP<0.05, dP<0.0001; two-tailed Student’s t-test.
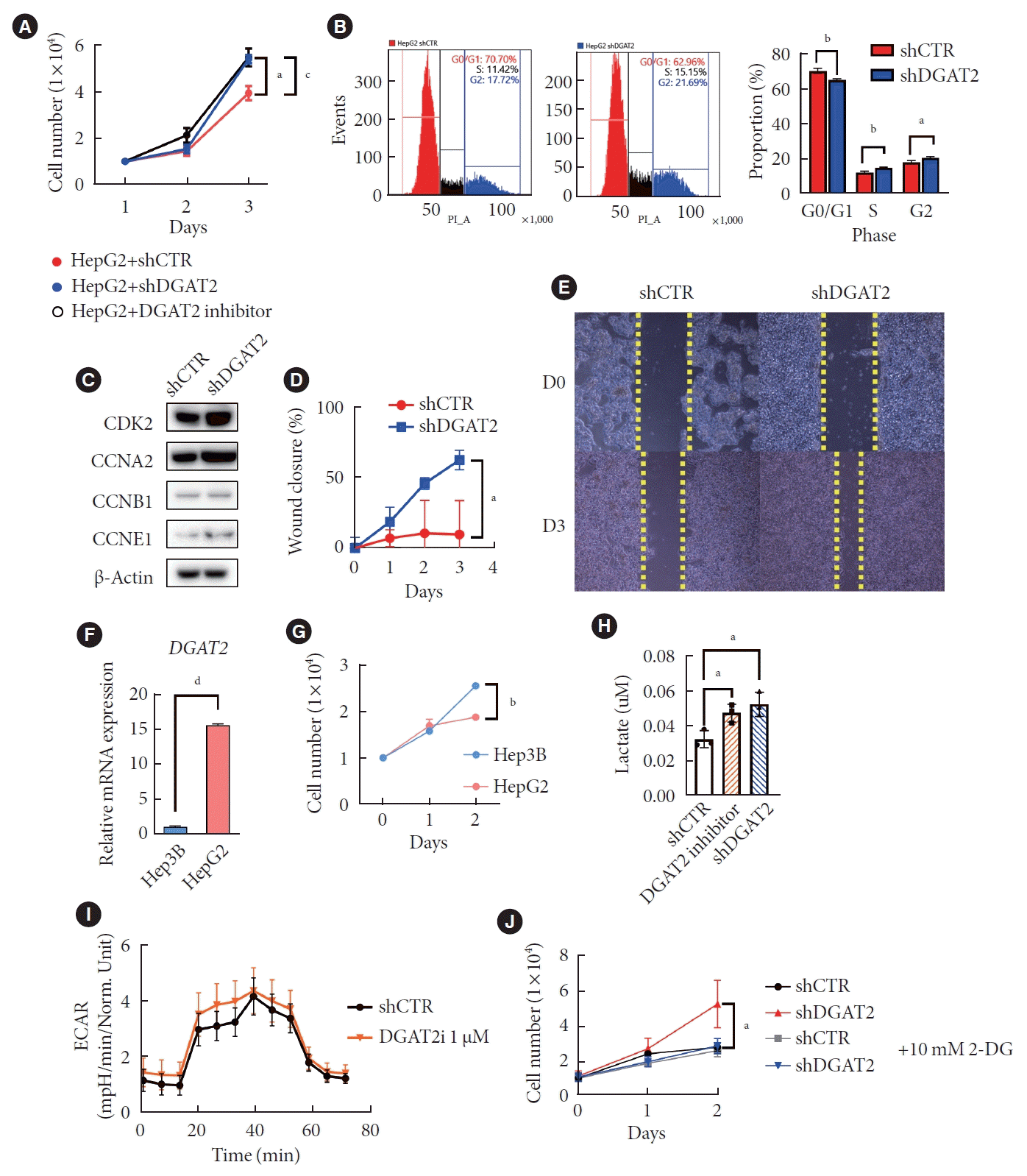
Fig. 5.
Estrogen-related receptor alpha (ESRRA) activity is decreases as diacylglycerol O-acyltransferase 2 (DGAT2) expression suppressed. (A) Top three motifs with similar activity trends with DGAT2 expression level. (B) ESRRA motif activity after DGAT2 suppression. (C) Luciferase activity of estrogen-related receptor response element (ERRE) firefly luciferase. (D) Cellular cholesterol level of DGAT2 knockdown (KD) HepG2. (E) Number of peaks on a transcription start site of genes with ERRE motifs. (F) Heatmap of genes with ERRE motifs. (G) Subcellular localization of proteins after DGAT2 KD. (H) Immunoprecipitation of anti-ESRRA. Data represent mean±standard deviation. PLAGL1, PLAG1 like zinc finger 1; NAFL, non-alcoholic fatty liver; NASH, nonalcoholic steatohepatitis; KLF16, KLF transcription factor 16; SP2, Sp2 transcription factor; shCTR, control shRNA; shDGAT2, DGAT2 shRNA; RLU, relative light unit; NS, not significant; PROX1, prospero homeobox 1; IgG, immunoglobulin G. aP<0.0001; two-tailed Student’s t-test.
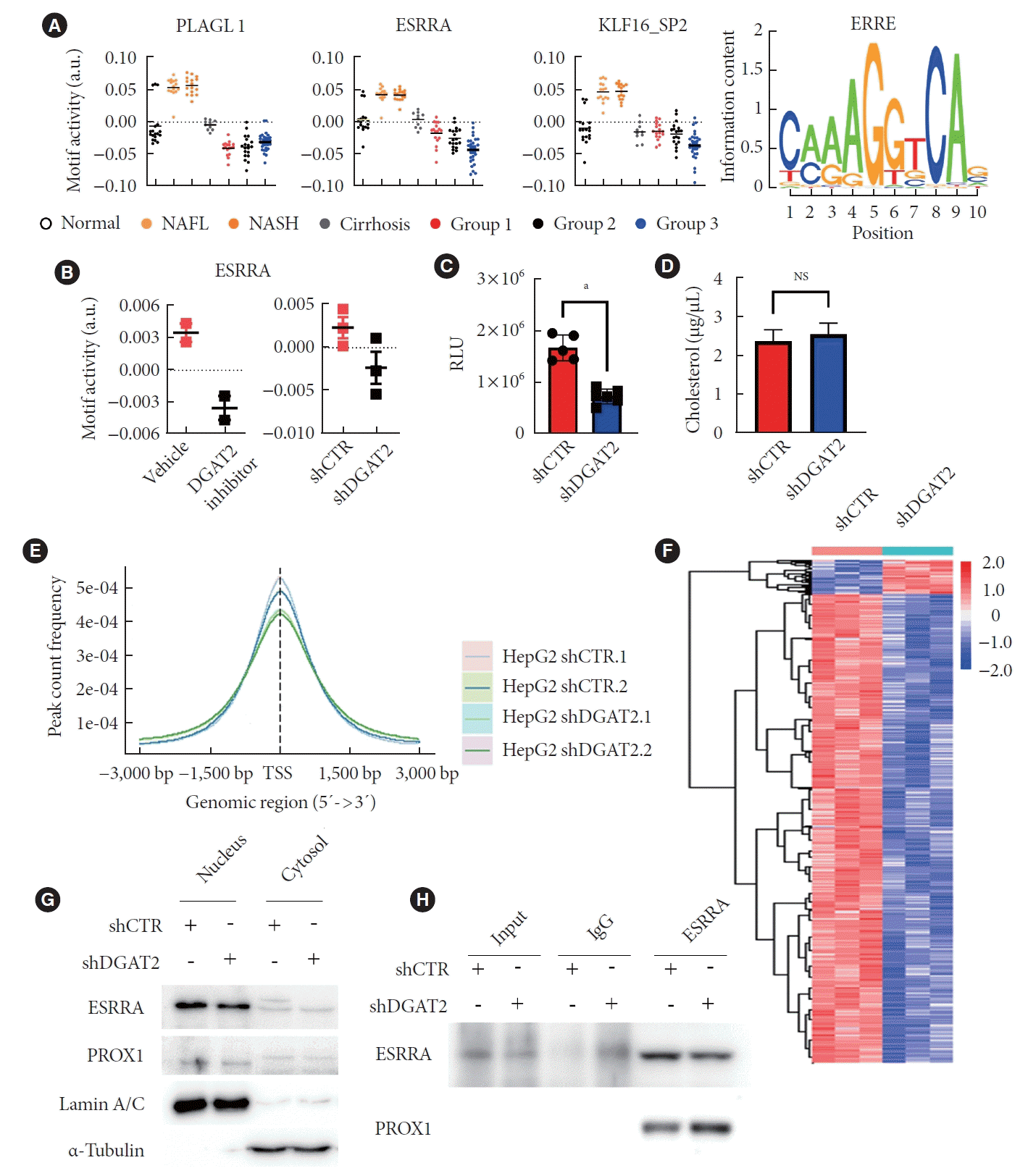
Fig. 6.
Overexpression of estrogen-related receptor alpha (ESRRA) rescues mitochondria dysfunction. (A) Transmission electron microscopy of rescued HepG2 exhibits mitochondrial recovery. (B) Relative mitochondrial area after diacylglycerol O-acyltransferase 2 (DGAT2) and ESRRA overexpression. (C) Mitochondrial length after gene overexpression. (D) Tetramethyl rhodamine ethyl ester (TMRE) intensity of HepG2 recovers after ESRRA overexpression. (E) Oxygen consumption rate (OCR) of ESRRA overexpressed HepG2. (F) Cell proliferation is suppressed after ESRRA overexpression. Data represent mean±standard deviation. shCTR, control shRNA; shDGAT2, DGAT2 shRNA; OE, overexpression; NS, not significant. aP<0.04, bP<0.01, cP<0.001, dP< 0.0001; two-tailed Student’s t-test.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download