Abstract
Purpose
Materials and Methods
Results
Electronic Supplementary Material
Notes
Ethical Statement
All patients provided written informed consent, and the study protocol was approved by the institutional review boards of the National Cancer Center Korea and Samsung Medical Center (NCC IRB # 2017-0127, 2019-0004, SMC IRB# 2014-11-015).
Author Contributions
Conceived and designed the analysis: Park KH, Park JY, Lee ES, Park YH, Kong SY.
Collected the data: Park KH, Park YH, Kong SY.
Contributed data or analysis tools: Kim CM, Park KH, Yu YS, Park YH, Kong SY.
Performed the analysis: Kim CM, Park KH, Yu YS, Kim JW, Park K, Yu JH, Lee JE, Park YH, Kong SY.
Wrote the paper: Kim CM, Park KH, Yu YS, Sim SH, Seo BK, Kim JK, Park YH, Kong SY.
Obtained funding: Park KH, Park JY, Park YH, Kong SY.
Administrative, technical, or material support: Park K, Park KH, Park YH, Lee ES, Kong SY, Park JY.
Conflicts of Interest
Chang Min Kim; Employee for CbsBioscience. Kyong Hwa Park; Consultancy/advisory role for: AstraZeneca, Pfizer, Eisai, Roche, Daiichi-Sankyo, Eli Lilly, and Novartis Pharmaceuticals; Honoraria from: AstraZeneca, Pfizer, Eisai, Roche, Daiichi-Sankyo, iMBDx, and Novartis; Grant/Research funding from AstraZeneca. Yun Suk Yu; Employee for CbsBioscience. Jin Young Park; Employer of CbsBioscience. Yeon Hee Park; Consultancy/advisory role for: AstraZeneca, Pfizer, Eisai, Roche, Daiichi-Sankyo, Eli Lilly, and Novartis Pharmaceuticals; Patents and royalties from: Hanmi; Honoraria from: AstraZeneca, Pfizer, Eisai, Roche, Daiichi-Sankyo, and Novartis; Grant/Research funding from AstraZeneca, Merck, Pfizer, Novartis, Alteogen, and Roche. Sun-Young Kong; Grant/Research funding from GSK and iMBDx. All remaining authors have declared no conflicts of interest.
ACKNOWLEDGMENTS
References
Fig. 1.
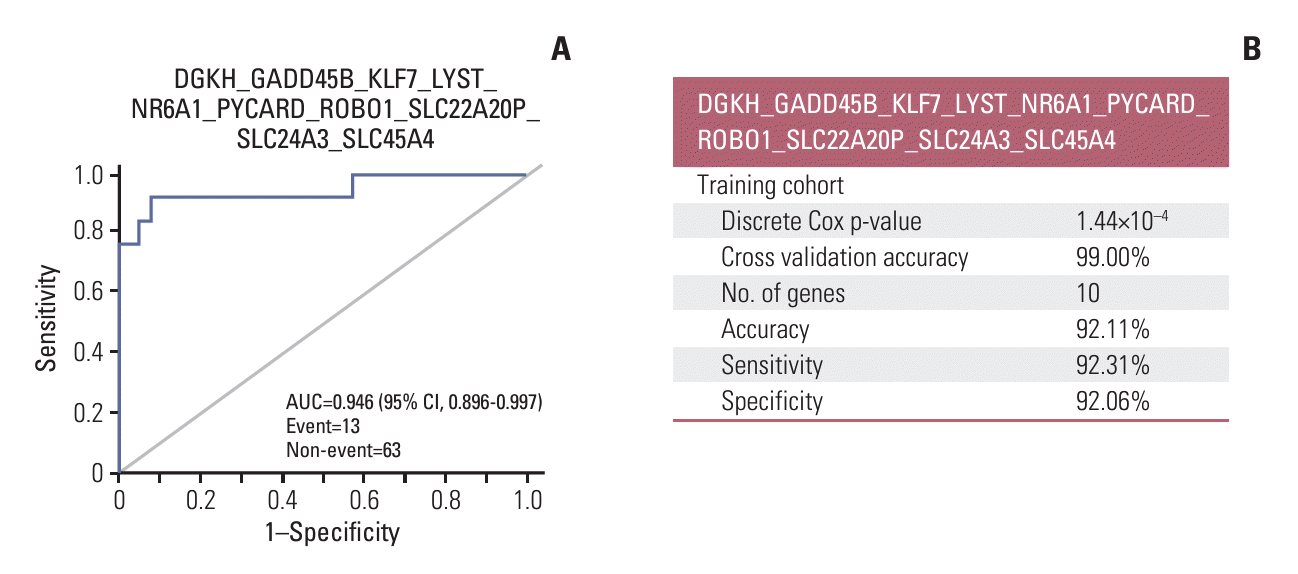
Fig. 2.
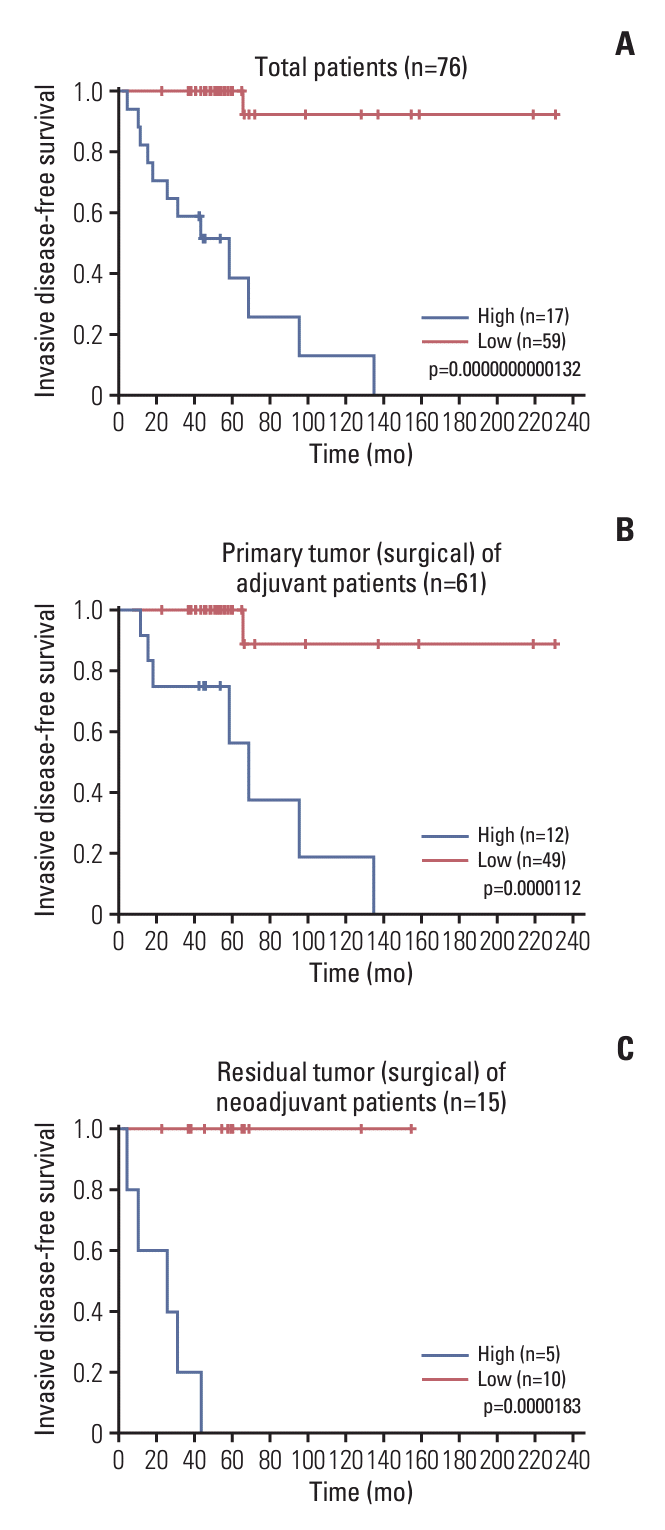
Fig. 3.
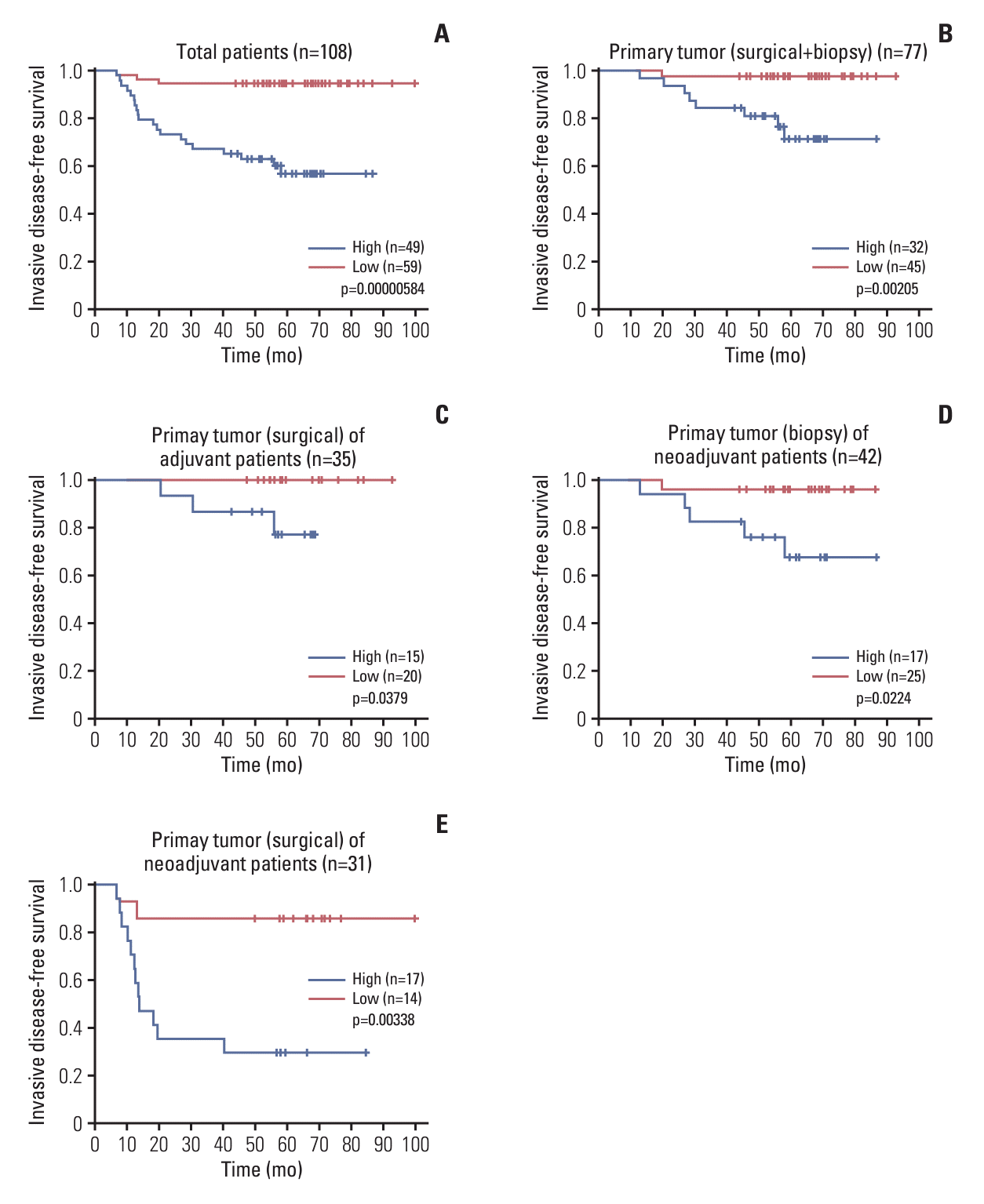
Fig. 4.
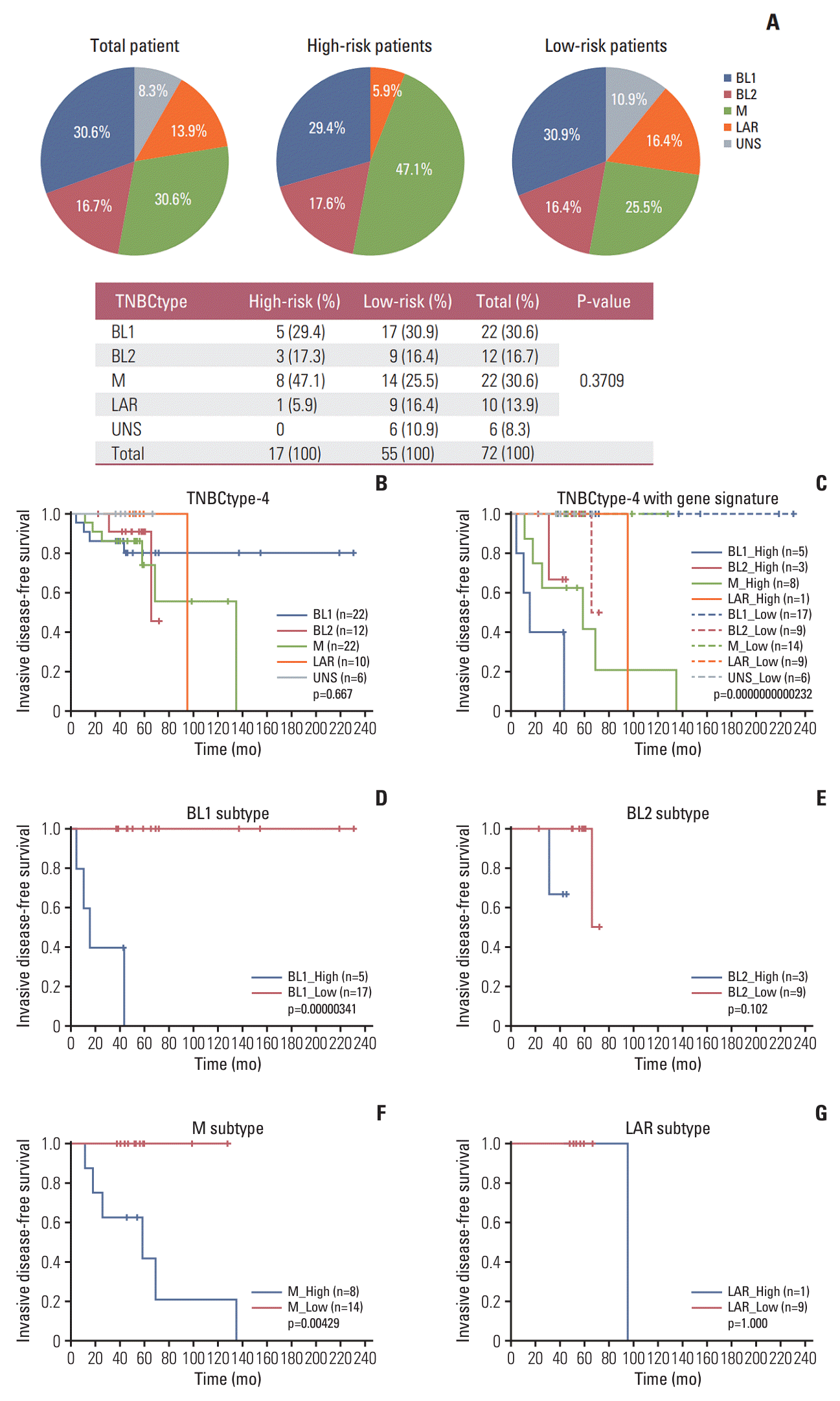
Table 1.
| Pathologic parameter |
Training cohort |
Validation cohort |
Training vs. validation p-valuea) | ||||
|---|---|---|---|---|---|---|---|
| Total (n=76) | Primary tumors (n=61) | Residual tumors (n=15) | Total (n=108) | Primary tumors (n=77) | Residual tumors (n=31) | ||
| Age (yr) | |||||||
| < 35 | 17 (22.4) | 10 (16.4) | 7 (46.7) | 36 (33.3) | 24 (31.2) | 12 (38.7) | 0.1465 |
| ≥ 35 | 59 (77.6) | 51 (83.6) | 8 (53.3) | 72 (66.7) | 53 (68.8) | 19 (61.3) | |
| TNM stage (pathologic) | |||||||
| I | 29 (38.2) | 29 (47.5) | - | 18 (16.7) | 18 (23.4) | - | 3.48×10–5 |
| II | 42 (55.3) | 32 (52.5) | 10 (66.7) | 57 (52.8) | 44 (57.1) | 13 (41.9) | |
| III | 5 (6.6) | - | 5 (33.3) | 33 (30.6) | 15 (19.5) | 18 (58.1) | |
| Systemic chemotherapy | |||||||
| Neo-adjuvant | 15 (19.7) | - | 15 (100) | 73 (67.6) | 42 (54.5) | 31 (100) | - |
| AC | 2 (13.3) | - | 2 (13.3) | 2 (2.7) | - | 2 (6.5) | |
| AC-D | 6 (40.0) | - | 6 (40.0) | 64 (87.7) | 35 (83.3) | 29 (93.5) | |
| AC-wP | - | - | - | 7 (9.6) | 7 (16.7) | - | |
| AC-PC | 4 (26.7) | - | 4 (26.7) | - | - | - | |
| PCarbo | 2 (13.3) | - | 2 (13.3) | - | - | - | |
| DA | 1 (6.7) | - | 1 (6.7) | - | - | - | |
| Adjuvant | 61 (80.3) | 61 (100) | - | 35 (32.4) | 35 (45.5) | - | |
| AC | 6 (9.8) | 6 (9.8) | - | 2 (5.7) | 2 (5.7) | - | |
| TC | 14 (23.0) | 14 (23.0) | - | 1 (2.9) | 1 (2.9) | - | |
| FAC | 14 (23.0) | 14 (23.0) | - | 3 (8.6) | 3 (8.6) | - | |
| AC-D | 7 (11.5) | 7 (11.5) | - | 5 (14.3) | 5 (14.3) | - | |
| AC-wP | 6 (9.8) | 6 (9.8) | - | 4 (11.4) | 4 (11.4) | - | |
| AC-PC | 3 (4.9) | 3 (4.9) | - | - | - | - | |
| TAC | 11 (18.0) | 11 (18.0) | - | 20 (57.1) | 20 (57.1) | - | |
| Event | |||||||
| Presence | 13 (17.1) | 8 (13.1) | 5 (33.3) | 23 (21.3) | 9 (11.7) | 14 (45.2) | 0.4057 |
| Absence | 63 (82.9) | 53 (86.9) | 10 (66.7) | 85 (78.7) | 68 (88.3) | 17 (54.8) | |
| Specimen | |||||||
| Core biopsy | - | - | - | 42 (38.9) | 42 (54.5) | - | |
| Surgery | 76 (100) | 61 (100) | 15 (100) | 66 (61.1) | 35 (45.5) | 31 (100) | |
| Follow-up period (mo), median (range) | 51.5 (4.6-230.8) | 51.1 (11.5-230.8) | 57.8 (4.6-154.8) | 58.3 (6.6-99.8) | 59.0 (12.9-92.7) | 56.6 (6.6-99.8) | |
Values are presented as number (%) unless otherwise indicated. AC, adriamycin, cyclophosphamide; AC-D, AC followed by docetaxel; AC-PC, AC followed by paclitaxel and carboplatin; AC-wP, AC followed by weekly paclitaxel; DA, docetaxel and adriamycin; Event, recurrence or metastasis; FAC, 5-FU, adriamycin, cyclophosphamide; PCarbo, paclitaxel and carboplatin; TAC, taxotere, and AC; TC, taxotere and cyclophosphamide; TNM, tumor-node-metastasis (American Joint Committee on Cancer stage).
Table 2.
AUC, area under the curve; CUEDC1, CUE domain containing 1; DGKH, diacylglycerol kinase eta; DCLK2, doublecortin like kinase 2; DIP2B, disco interacting protein 2 homolog B; EMP1, epithelial membrane protein 1; GADD45B, growth arrest and DNA damage inducible beta; KLF7, Kruppel-like factor 7; LYST, lysosomal trafficking regulator; MT2A, metallothionein 2A; NOTCH2, notch receptor 2; NOXA1, NADPH oxidase activator 1; NR6A1, nuclear receptor subfamily 6 group A member 1; NTAQ1, N-terminal glutamine amidase 1; PRICKLE1, prickle planar cell polarity protein 1; PYCARD, PYD and CARD domain containing; ROBO1, roundabout guidance receptor 1; RORA, RAR related orphan receptor A; SLC22A20P, solute carrier family 22 member 20, pseudogene; SLC24A3, solute carrier family 24 member 3; SLC45A4, solute carrier family 45 member 4; SLC6A20, solute carrier family 6 member 20.
Table 3.
CI, confidence interval; DGKH, diacylglycerol kinase eta; GADD45B, growth arrest and DNA damage inducible beta; HR, hazard ratio; KLF7, Kruppel-like factor 7; LYST, lysosomal trafficking regulator; NR6A1, nuclear receptor subfamily 6 group A member 1; PYCARD, PYD and CARD domain containing; RC, regression coefficient; ROBO1, roundabout guidance receptor 1; ROR-S, risk of recurrence based on subtype; SLC22A20P, solute carrier family 22 member 20, pseudogene; SLC24A3, solute carrier family 24 member 3; SLC45A4, solute carrier family 45 member 4; TCRβ, T cell receptor beta locus; TNM, tumor-node-metastasis (American Joint Committee on Cancer stage).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download