Abstract
Background
In the early days of the coronavirus disease 2019 (COVID-19) pandemic, unclear discharge guidelines for hospitalized COVID-19 patients resulted in numerous individuals being kept in the hospital until their real-time reverse transcription-polymerase chain reaction (rRT-PCR) results showed negativity. The objective of this study was to analyze the variations in repeated follow-up rRT-PCR results among hospitalized patients.
Methods
We analyzed rRT-PCR results from 84 patients diagnosed with COVID-19 at Daegu Catholic University Medical Center in 2020. The PowerChek 2019-nCoV Real-time PCR Kit (KogeneBiotech, Korea) and CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, USA) were used, and the delta cycle threshold (ΔCt) values were retrospectively analyzed alongside the clinical features.
Results
The patients underwent nine repeat rRT-PCR tests over a span of 38.5 days, showing negative results after an average of 28.0 days. The median number of days required to achieve the first negative result was 27.5 days for the consistently negative group and 37.0 days for the negative-positive-negative (re-positive) group. The ΔCt values for the E and RdRp genes increased continuously from their initial values on the day of hospitalization, peaked at 25.0 days post-hospitalization, and then decreased thereafter.
Conclusions
In contrast to the patients in the consistently negative group, those in the re-positive group took an additional 10.0 days to achieve SARS-CoV-2 negativity in the rRT-PCR test. Therefore, repeated rRT-PCR tests after 4 weeks may not accurately reflect the actual viral load of clinically stable patients.
초록
배경
코로나 유행 초기에는 COVID-19로 입원한 환자에 대한 퇴원 지침이 명확하지 않아서 많은 환자들이 SARS-COV-2 real-time reverse transcription-PCR (rRT-PCR) 결과가 음성으로 나올 때까지 퇴원이 보류되었다. 저자들은 COVID-19 환자에서 연속적으로 시행한 rRT-PCR 검사 결과의 변화 양상을 분석하고자 한다.
방법
2020년 대구가톨릭대학교병원에서 COVID-19로 진단받은 84명 환자의 rRT-PCR 결과를 분석하였다. PowerCheck 2019-nCoV 키트(KogeneBiotech, Korea)와 CFX 96 Real-Time System (Bio-Rad Laboratories, USA)이 사용되었으며, PCR의 델타 Ct 값과 임상 특징을 후향적으로 분석하였다.
In December 2019, during the early stages of the coronavirus disease 2019 (COVID-19) outbreak, discharge guidelines for hospitalized patients were not clear, leading to many patients remaining hospitalized until their real-time reverse transcription-polymerase chain reaction (rRT-PCR; hereinafter referred to as PCR) tests returned negative results. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PCR test is crucial for diagnosing COVID-19 [1, 2], monitoring the disease progression [3, 4], and determining whether hospitalized patients can be discharged [5, 6]. However, interpreting the PCR result can be more challenging during follow-up than at the time of diagnosis. During the initial diagnosis, a large amount of virus particles is present in the test specimen, making it easy to determine positive or negative results. However, during follow-up testing, especially at the time of discharge, the virus concentration is low, rendering interpretation of the results difficult.
Since the emergence of COVID-19 in 2019, numerous studies have reported on the diagnosis [7, 8], clinical features [9, 10], and treatment of the disease [11, 12]. Additionally, the implementation and clinical application of the PCR-based SARS-CoV-2 detection assay have been well reported [13-15]. However, few studies have conducted follow-up PCR testing on SARS-CoV-2-positive patients and analyzed the results. Therefore, the aim of our present study was to analyze changes in the follow-up PCR results obtained from the repeated testing of hospitalized COVID-19 patients.
The diagnostic and follow-up data used in this study were from COVID-19 patients in Daegu, the site of the first large-scale infection in Korea. During the study period, COVID-19 diagnosis was conducted using the SARS-CoV-2 rRT-PCR assay, making analysis of the results challenging. There were instances where it was difficult to determine the results at the time of monitoring and discharge. By analyzing the follow-up PCR results of SARS-CoV-2-positive patients in detail, we aim to provide useful information that can be used to improve the diagnosis and monitoring of COVID-19 or any new infectious diseases.
This study was approved by the Institutional Review Board of Daegu Catholic University Medical Center. We analyzed the PCR results of patients diagnosed with COVID-19 at Daegu Catholic University Medical Center from March to May 2020. In total, 84 patients who underwent four or more PCR tests and had their medical records confirmed were selected for the study. The PowerCheck 2019-nCoV Kit (Kogene Biotech, Seoul, Korea) and CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) were used for the SARS-CoV-2 PCR tests, and delta cycle threshold (ΔCt) values and clinical features were retrospectively analyzed.
A Ct value of 39 was used as the cut-off for determining positive and negative PCR results. PCR negativity was defined as the first days when negative results were received twice in a row [16]. If either the E or RdRp gene alone was positive, the results were considered positive when the positive results persisted and negative when the negative results persisted. For patients who tested negative, a subsequent positive and then negative result was determined on the basis of both the E and RdRp genes being positive and negative, respectively. To observe changes in the Ct values (i.e., ΔCt ≥8.0), the results for 20 patients with a PCR Ct value of 30 or less at the time of initial diagnosis were evaluated for changes based on the duration of hospitalization.
Statistical analyses were performed using MedCalc software (version 20.218; MedCalc Software Ltd., Ostend, Belgium). Oneway analysis of variance was used to analyze the differences in ΔCt values among the follow-up PCR subgroups. The Wilcoxon signed-rank test was used to compare the Ct values of the E and RdRp genes. The Mann–Whitney test was used to compare time intervals for negative conversion of the SARS-CoV-2 PCR results. A P-value of less than 0.05 was considered statistically significant.
Demographic data and SARS-Cov2 rRT-PCR results are summarized in Table 1. Of the 84 patients (median age, 67 years), 50 were females and 34 were males. At the time of initial diagnosis, their median Ct values for the E and RdRp genes were 33.3 and 33.6, respectively. These patients underwent nine repeat PCR tests over a span of 38.5 days, obtaining a negative result after a median of 28.0 days. Among the 84 patients, 77 tested negative and 7 remained PCR positive throughout the study period (Fig. 1). Of the 77 patients with negative results, 58 remained negative (consistently negative group), while 19 later tested positive before subsequently returning to a negative status (re-positive group). The Ct values of the E and RdRp genes at the time of re-positivity were 34.7 and 35.2, respectively, which were higher than the values recorded at the initial diagnosis (Table 1).
Table 2 presents the median ΔCt values for the E and RdRp genes based on the duration of hospitalization. The ΔCt value was calculated by subtracting the present Ct value from the initial value obtained on the duration of hospitalization. The ΔCt values of the two genes showed a continuous increase from 5.16 to 12.6 (E gene) and from 5.15 to 12.02 (RdRp gene) until the 25th day of hospitalization, indicating a decline in the gene concentrations over time. Notably, however, the ΔCt values of both genes started to decrease after the 25th day.
The concordance rate of the PCR test results for the E and RdRp genes was 82.4%, with both genes being positive in 424 samples (60.2%) and negative in 156 samples (22.2%) (Table 3). However, the PCR test discrepancy rate between the two genes was 17.6%, with 8.1% of samples testing positive for the E gene and negative for the RdRP gene and 9.5% testing positive for the RdRp gene and negative for the E gene.
Among the 424 positive samples, the median Ct values of the E and RdRp genes were 34.43 and 35.07, respectively, with the higher Ct value for the RdRP gene being statistically significant (P<0.001) (Fig. 2).
The time interval for negative PCR conversion was compared between the consistently negative and re-positive groups. The mean number of negative conversion days for the negative and re-positive groups were 27.8 and 33.7 days, respectively, with the longer period for the re-positive group being statistically significant (P<0.05) (Fig. 3).
Of the 84 patients in the study, seven tested persistently positive for SARS-CoV-2. Among these seven patients, three succumbed to the illness, one required transfer to an intensive care center, and three showed improvement and were subsequently transferred to an isolated observation center (Table 4). In the four patients with a poor prognosis, the levels of C-reactive protein (CRP) remained elevated. Conversely, in the three patients who showed improvement, the CRP levels decreased significantly. Notably, among the four patients with initial Ct values of approximately 25 (Patients 2, 4, 6, and 7), the two with a poor prognosis exhibited only a slight change in Ct values (2.5–3.9 cycles for patients 4 and 7), whereas the other two with a favorable prognosis showed a substantial change in Ct values (10–12.2 cycles for patients 2 and 6).
To address the lack of data on the ideal frequency and duration of PCR testing for COVID-19 patients, we analyzed the follow-up PCR records of 84 SARS-CoV-2-positive patients to evaluate the optimal duration and frequency of repeat testing. Challenges include discrepancies between the PCR results of the two genes tested and difficulties in adhering to the cut-off Ct values for positive results as suggested by manufacturers.
In this study, in which a total of nine tests were performed at intervals of 4.3 days over a period of 38.5 days, the median duration required for negative PCR conversion was 28.0 days. The median duration of SARS-CoV-2 detection in the upper respiratory tract has been reported to be 14.5–34.0 days [16-19]. Generally, the viral load of SARS-CoV-2 is very high on the day of symptom development and then gradually decreases, with the virus typically being undetectable within 3 weeks of symptom onset [16, 17, 20]. The duration of virus detection also depends on the chosen cutoff Ct value [19].
In the ΔCt analysis based on the duration of hospitalization, the viral concentration decreased rapidly until the second week of hospitalization and then decreased gradually thereafter, but it then surprisingly increased again after the fourth week. In clinically stable patients, the increase in viral concentrations after the fourth week does not indicate active viral replication but rather the presence of residual nucleic acid or persistent inactive viral particles. Therefore, repeated PCR testing for SARS-CoV-2 after 4 weeks may not accurately reflect the actual viral load in the patient.
Among the 77 patients who tested negative for SARS-CoV-2, 19 (22.6%) experienced PCR re-positivity (Fig. 1). Moreover, the median duration of the first virus-negative period was significantly longer in the re-positive group than in the consistently negative group (37.0 vs. 27.5 days). Wong et al. [21] reported that 34.3% of 35 patients showed SARS-CoV-2 re-positivity, without COVID-19 symptoms, likely as a result of residual viral nucleic acid. However, Hossain et al. [18] found that two of 12 patients with symptom recurrence also had a re-positive PCR result. Intermittent viral shedding has also been identified as a major cause of PCR re-positivity [22]. Hu et al. [23] reported that delayed clearance of SARSCoV-2 RNA was associated with an older age and chest tightness in hospitalized patients, whereas Abrahim et al. [24] found that patients with comorbidities, such as cancer and HIV infection, had a prolonged viral clearance period. Lim et al. [25] suggested that the duration of viral shedding was longer in individuals with more clinical symptoms, indicating that the time required for negative conversion might be related to the severity of clinical symptoms.
Compared with the 58 consistently virus-negative patients, the 19 re-positive patients were significantly older (median age 61.5 vs. 81.0 years), whereas other clinical symptoms were generally unremarkable. Among the 19 re-positive patients, 12 had CRP levels within the normal range, and two showed slightly increased levels (5.0–12.0 mg/dL). Seventeen of the re-positive patients recovered from COVID-19, whereas the remaining two patients experienced severe respiratory symptoms and died from respiratory failure. The re-positivity in the 17 recovered patients was likely due to residual nucleic acid or intermittent viral shedding. By contrast, the re-positivity in the two deceased patients may have been due to delayed viral clearance or persistent viral presence, indicating the potential ongoing complications of COVID-19.
The Ct values of the RdRp gene were higher than those of the E gene, aligning with the findings from another study that used the same PCR reagent [13]. This discrepancy in Ct values is attributed to variations in PCR conditions rather than differences in target gene concentration. Furthermore, the PCR concordance rate between the two genes was 82.4%, reflecting a mismatch rate of 17.6% (Table 3). Sung et al. [13] reported that in follow-up PCRs of confirmed infected patients, only one of the target genes might test positive and the detected gene may vary.
Of the seven patients who remained persistently virus positive, three experienced symptom improvement, whereas the other four individuals either died or required intensive care. In a study comparing 67 patients, Chang et al. [4] found that patients who tested persistently positive for SARS-CoV-2 had higher incidences of intensive care requirement and pulmonary infiltration. Conversely, Ikegami et al. [20] reported that six out of 272 recovered patients continued to test positive for SARS-CoV-2 in nasopharyngeal swab PCR assays. This indicates that persistent viral detection can occur in both recovered and deteriorating patients. Therefore, the clinical significance of PCR results has to be interpreted in conjunction with CRP levels or ΔCt values.
In summary, it took a median of 28.0 days for 77 patients to test negative in the SARS-CoV-2 PCR assay, with the re-positive group taking a median of 37.0 days to achieve this. An 82.4% concordance rate between the E and RdRp genes was observed. We conclude that for clinically stable patients, continued follow-up PCR testing for up to 4 weeks is advisable to assess negative PCR conversion.
REFERENCES
1. Machado BAS, Hodel KVS, Barbosa-Júnior VG, Soares MBP, Badaró R. 2020; The main molecular and serological methods for diagnosing COVID-19: an overview based on the literature. Viruses. 13:40. DOI: 10.3390/v13010040. PMID: 0e2f4541f6e44ffea3a89b9652031e36.
2. Younes N, Al-Sadeq DW, Al-Jighefee H, Younes S, Al-Jamal O, Daas HI, et al. 2020; Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 12:582. DOI: 10.3390/v12060582. PMID: b775107b8e9b4c3d93b48a9001ba40d1.
3. Aranha C, Patel V, Bhor V, Gogoi D. 2021; Cycle threshold values in RT-PCR to determine dynamics of SARS-CoV-2 viral load: an approach to reduce the isolation period for COVID-19 patients. J Med Virol. 93:6794–7. DOI: 10.1002/jmv.27206. PMID: 34264527. PMCID: PMC8426941.
4. Chang D, Zhao P, Zhang D, Dong JH, Xu Z, Yang G, et al. 2020; Persistent viral presence determines the clinical course of the disease in COVID-19. J Allergy Clin Immunol Pract. 8:2585–91.e1. DOI: 10.1016/j.jaip.2020.06.015. PMID: 32574840. PMCID: PMC7305869.
5. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. 2020; Virological assessment of hospitalized patients with COVID-2019. Nature. 581:465–9. DOI: 10.1038/s41586-020-2196-x. PMID: 32235945.
6. La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. 2020; Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 39:1059–61. DOI: 10.1007/s10096-020-03913-9. PMID: 32342252. PMCID: PMC7185831.
7. Binnicker MJ. 2020; Emergence of a novel coronavirus disease (COVID-19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 66:664–6. DOI: 10.1093/clinchem/hvaa071. PMID: 32077933. PMCID: PMC7108487.
8. Benzigar MR, Bhattacharjee R, Baharfar M, Liu G. 2021; Current methods for diagnosis of human coronaviruses: pros and cons. Anal Bioanal Chem. 413:2311–30. DOI: 10.1007/s00216-020-03046-0. PMID: 33219449. PMCID: PMC7679240.
9. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. 2020; Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 395:507–13. DOI: 10.1016/S0140-6736(20)30211-7.
10. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. 2020; Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. DOI: 10.1016/S0140-6736(20)30183-5. PMID: 31986264. PMCID: PMC7159299.
11. Majumder J, Minko T. 2021; Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 23:14. DOI: 10.1208/s12248-020-00532-2.
12. Fernandes Q, Inchakalody VP, Merhi M, Mestiri S, Taib N, Moustafa Abo El-Ella D, et al. 2022; Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann Med. 54:524–40. DOI: 10.1080/07853890.2022.2031274. PMID: 35132910. PMCID: PMC8843115. PMID: ee3ec5dd64c44c558549a2623b2047e9.
13. Sung H, Roh KH, Hong KH, Seong MW, Ryoo N, Kim HS, et al. 2020; COVID-19 molecular testing in Korea: practical essentials and answers from experts based on experiences of emergency use authorization assays. Ann Lab Med. 40:439–47. DOI: 10.3343/alm.2020.40.6.439. PMID: 32539299. PMCID: PMC7295959.
14. Garg A, Ghoshal U, Patel SS, Singh DV, Arya AK, Vasanth S, et al. 2021; Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. 93:2281–6. DOI: 10.1002/jmv.26691. PMID: 33230819. PMCID: PMC7753435.
15. Hong KH, Lee SW, Kim TS, Huh HJ, Lee J, Kim SY, et al. 2020; Guidelines for laboratory diagnosis of coronavirus disease 2019 (COVID-19) in Korea. Ann Lab Med. 40:351–60. DOI: 10.3343/alm.2020.40.5.351.
16. Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. 2020; SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 81:357–71. DOI: 10.1016/j.jinf.2020.06.067. PMID: 32615199. PMCID: PMC7323671.
17. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. 2021; SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2:e13–22. DOI: 10.1016/S2666-5247(20)30172-5. PMID: 33521734. PMCID: PMC7837230.
18. Hossain ME, Lister D, Bartolo C, Kinsella PM, Knox J, Aldrich R, et al. 2021; Prolonged viral shedding in patients with mild to moderate COVID-19 disease: a regional perspective. Infect Dis (Auckl). 14:11786337211010428. DOI: 10.1177/11786337211010428. PMID: 33911876. PMCID: PMC8047841.
19. Park SY, Yun SG, Shin JW, Lee BY, Son HJ, Lee S, et al. 2020; Persistent severe acute respiratory syndrome coronavirus 2 detection after resolution of coronavirus disease 2019-associated symptoms/signs. Korean J Intern Med. 35:793–6. DOI: 10.3904/kjim.2020.203. PMID: e0d2de1d6f5b4fd0bed4333c25907ff9.
20. Ikegami S, Benirschke R, Flanagan T, Tanna N, Klein T, Elue R, et al. 2020; Persistence of SARS-CoV-2 nasopharyngeal swab PCR positivity in COVID-19 convalescent plasma donors. Transfusion. 60:2962–8. DOI: 10.1111/trf.16015. PMID: 32840002. PMCID: PMC7461313.
21. Wong CL, Lei SK, Lei CI, Lo IL, Lam C, Leong IH. 2021; Re-positive of SARSCoV-2 test is common in COVID-19 patients after hospital discharge. Data from high standard post-discharge quarantined patients in Macao SAR, China. PeerJ. 9:e11170. DOI: 10.7717/peerj.11170. PMID: 47d3c454b60848d28eab7555aea39739.
22. Dao TL, Hoang VT, Gautret P. 2021; Recurrence of SARS-CoV-2 viral RNA in recovered COVID-19 patients: a narrative review. Eur J Clin Microbiol Infect Dis. 40:13–25. DOI: 10.1007/s10096-020-04088-z.
23. Hu X, Xing Y, Jia J, Ni W, Liang J, Zhao D, et al. 2020; Factors associated with negative conversion of viral RNA in patients hospitalized with COVID-19. Sci Total Environ. 728:138812. DOI: 10.1016/j.scitotenv.2020.138812. PMID: 32335406. PMCID: PMC7175870.
24. Abrahim SA, Tessema M, Defar A, Hussen A, Ejeta E, Demoz G, et al. 2020; Time to recovery and its predictors among adults hospitalized with COVID-19: a prospective cohort study in Ethiopia. PLoS One. 15:e0244269. DOI: 10.1371/journal.pone.0244269. PMID: 33378367. PMCID: PMC7773180. PMID: f897257f8d354c0bb75c762acbb80e62.
25. Lim AY, Cheong HK, Oh YJ, Lee JK, So JB, Kim HJ, et al. 2021; Modeling the early temporal dynamics of viral load in respiratory tract specimens of COVID-19 patients in Incheon, the Republic of Korea. Int J Infect Dis. 108:428–34. DOI: 10.1016/j.ijid.2021.05.062. PMID: 34058374. PMCID: PMC8161782.
Fig. 1
Time interval of SARS-CoV-2 rRT-PCR positivity status of 84 COVID-19 patients. Data are presented as median days (d).
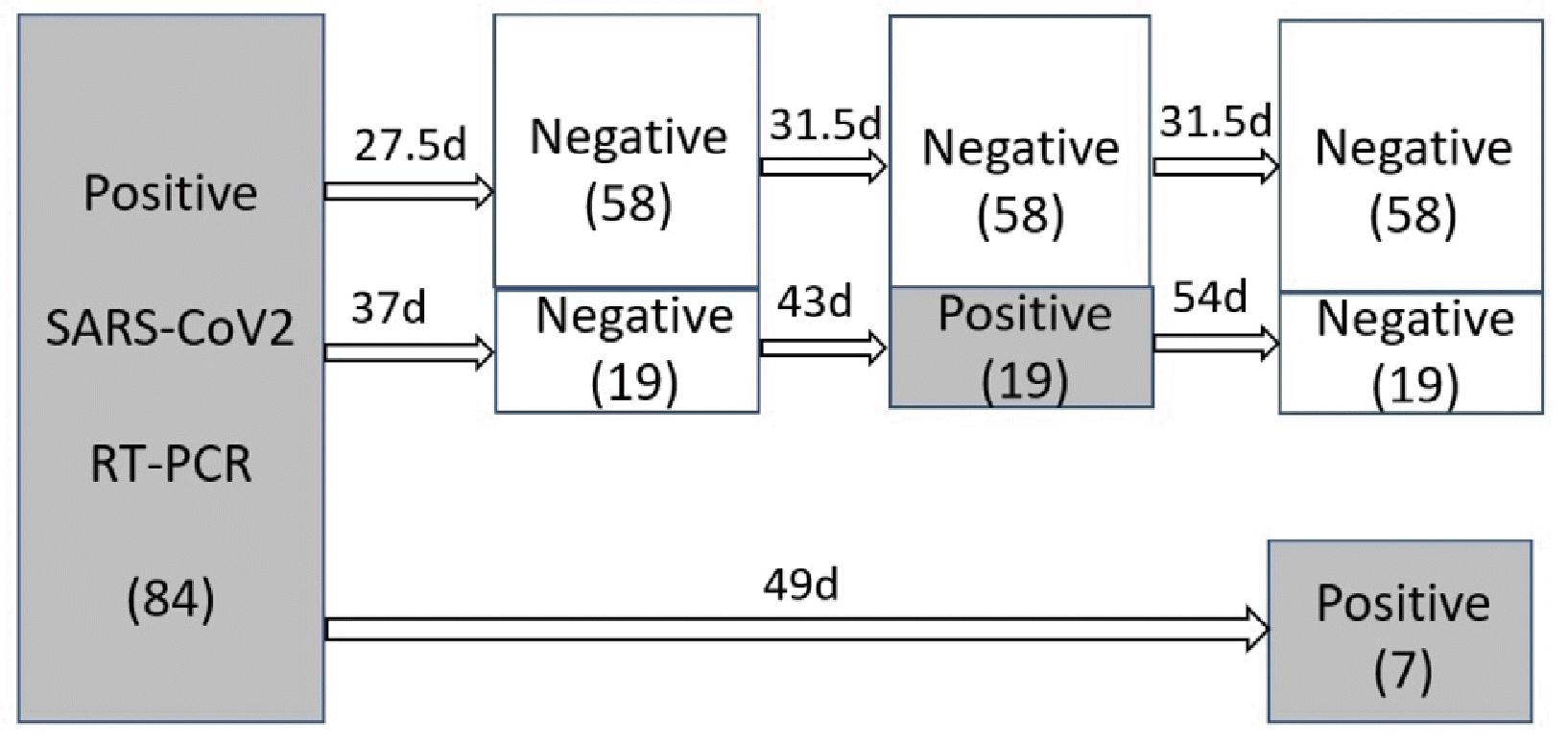
Fig. 2
Comparison of the Ct values (median and interquartile range) of the E and RdRp genes for the 424 SARS-CoV-2-positive samples (Box and whisker plot, P<0.0001 by the Wilcoxon signed-rank test).
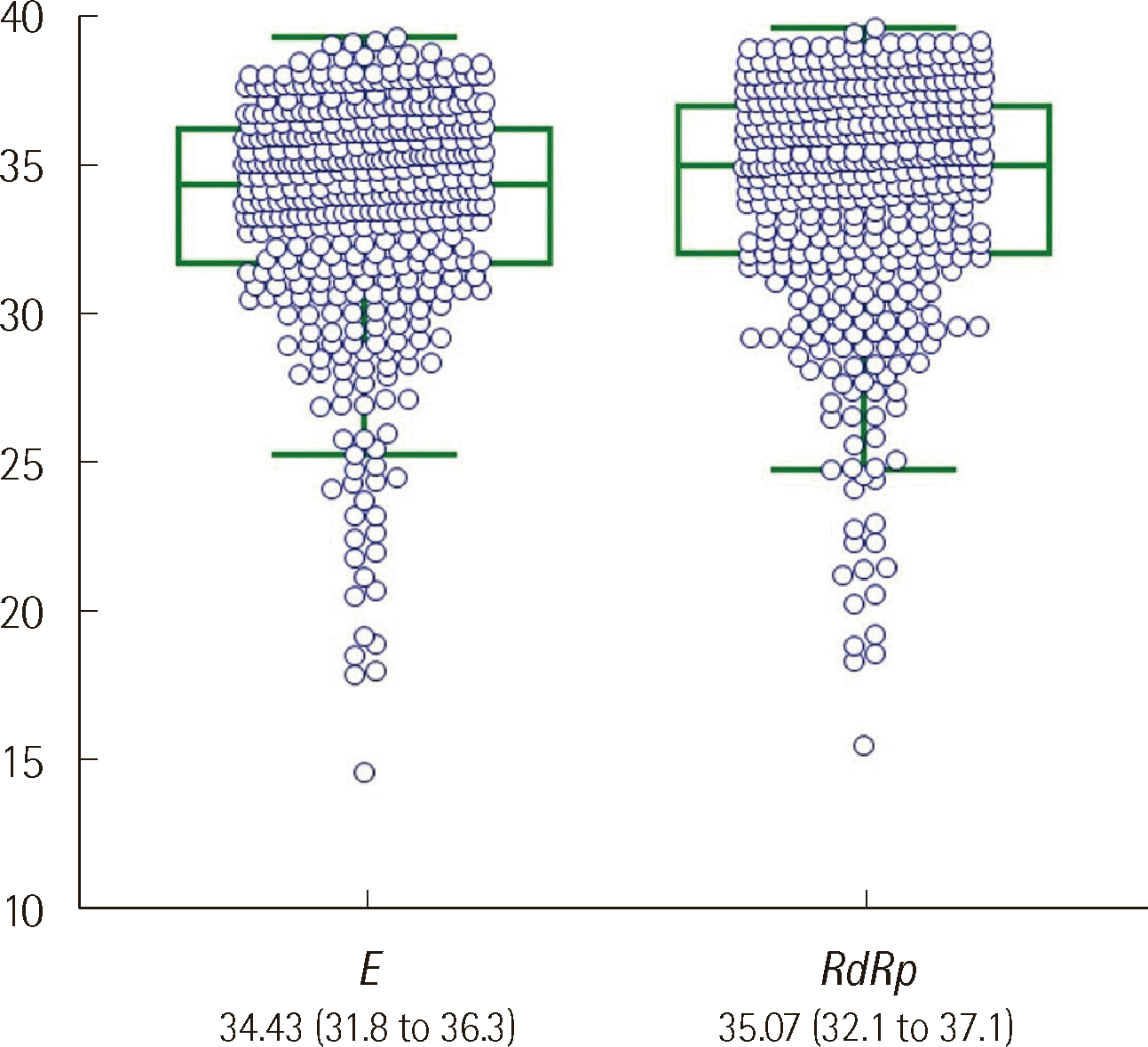
Fig. 3
Comparison of the time intervals (days, mean±2 SD) for negative SARS-CoV2 rRT-PCR conversion between the consistently negative and re-positive groups (P=0.049 by the independent t-test).
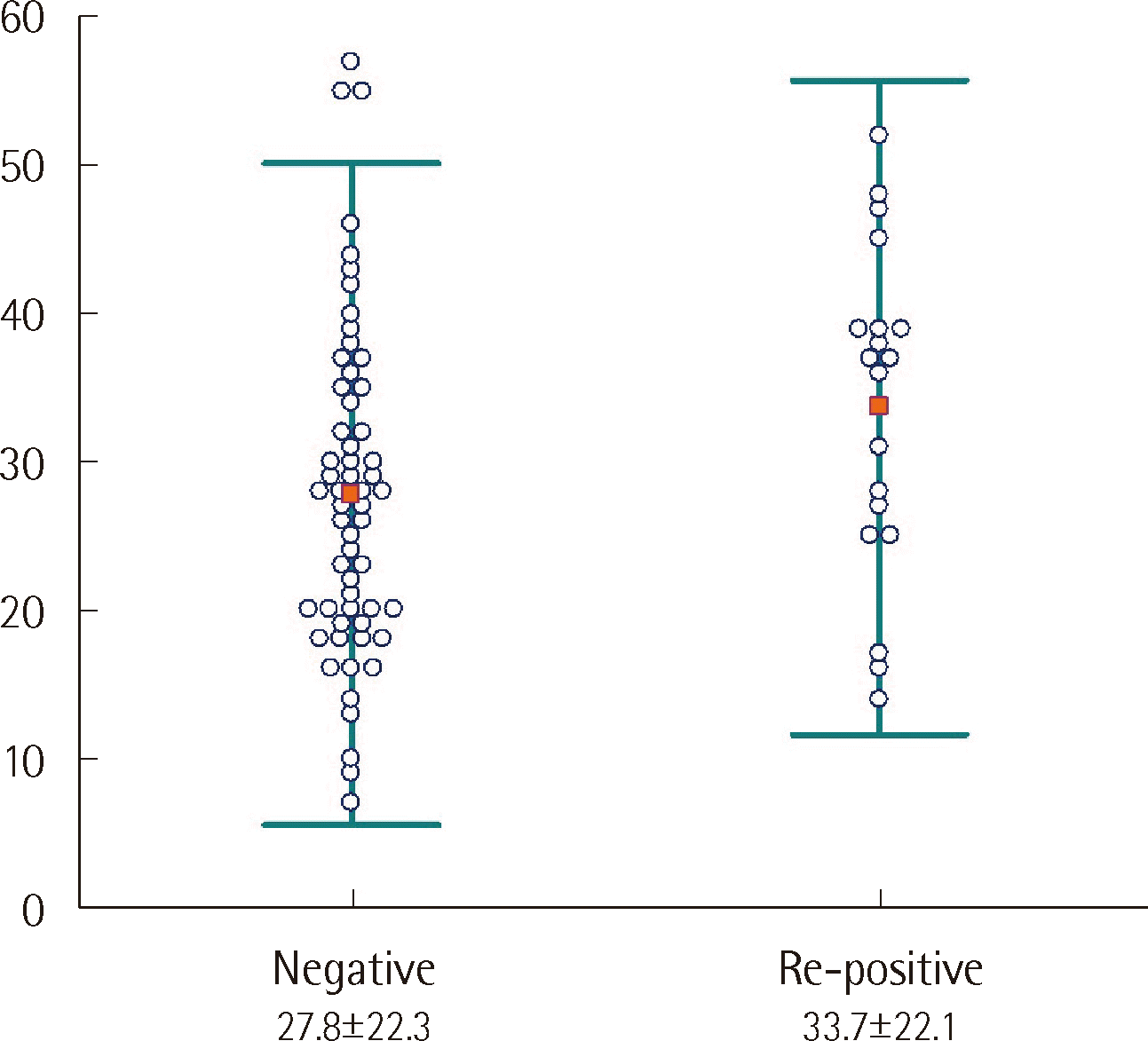
Table 1
Summary of follow-up SARS-CoV2 rRT-PCR results from repeated testing of hospitalized patients
Table 2
Median ΔCt values of the E and RdRp genes after the initial diagnosis
Table 3
Concordance rates of the rRT-PCR results for the E and RdRp genes
Table 4
Summary of rRT-PCR data and clinical outcomes of patients with persistent SARS-CoV2-positive results




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download