Abstract
Background
Pseudohypoparathyroidism type Ib (PHP-1b) is a rare imprinting disorder characterized by renal parathyroid hormone resistance, but without the physical features of Albright hereditary osteodystrophy. A common heterozygous 3-kb deletion of STX16 was previously reported in multiple unrelated probands. This deletion causes isolated loss of methylation at GNAS exons A/B. We aimed to identify STX16 deletions and the exact breakpoints to gain insights into the mechanism of STX16 deletion.
Methods
We investigated 10 patients who were diagnosed with pseudohypoparathyroidism type Ib, but did not have any pathogenic variants in GNAS. A methylation-specific multiplex ligation-dependent probe amplification assay of STX16 was performed to assess allelic dosage. Junction PCR and Sanger sequencing were performed around the presumptive breakpoint area to determine the exact breakpoint.
Results
STX16 gene deletion was detected in two of the 10 probands (20%). The deletion range was the same for both probands, ranging from g.57,243,566 to g.57,246,545 (2,979 bp), according to the February 2009 human reference sequence (hg19, build37), which is consistent with previous reports. The 5’ and 3’ break points were located in the mammalian-wide interspersed repeats (MIRs) sequences suggesting MIR-mediated non-allelic homologous recombination as the putative mechanism for the common STX16 3-kb deletion.
초록
배경
가성부갑상선기능저하증 1b형(PHP-1b)은 드문 각인 장애로, 신장에서 부갑상선 호르몬 저항성을 특징으로 하지만, Albright 유전성 골이영양증의 신체적 특징은 나타나지 않는다. 이전에 보고된 바에 따르면, 여러 비관련 환자들에서 공통적으로 나타나는 이형접합성 3kb STX16 결실이 있다. 이 결실은 GNAS 유전자 A/B 엑손에서 메틸화 문제를 유발한다. 저자들은 STX16 결실과 정확한 절단점을 확인하여 STX16 결실의 메커니즘을 규명하고자 하였다.
방법
저자들은 PHP-1b로 진단받았으나 GNAS 유전자에 병원성 서열 변이가 없는 10명의 환자를 대상으로 연구하였다. STX16의 복제수를 평가하기 위해 메틸화 특이적 복합결찰 의존 탐침 증폭법(methylation-specific multiplex ligation-dependent probe amplification, MS-MLPA) 분석을 수행하였다. 절단점 부근에서의 PCR과 생거 시퀀싱을 통해 정확한 절단점을 결정했다.
Disorders of GNAS inactivation include pseudohypoparathyroidism types Ia, Ib, and Ic (PHP-Ia, -Ib, and -Ic), pseudopseudohypoparathyroidism (PPHP), progressive osseous heteroplasia (POH), and osteoma cutis (OC). PHP-Ia and PHP-Ic feature hormone resistance and characteristics of Albright hereditary osteodystrophy (AHO), such as short stature and subcutaneous ossification. PHP-Ib is primarily involved in parathyroid hormone resistance. PPHP manifests as AHO without hormone resistance. POH involves progressive bone formation in the muscles and fascia, whereas OC involves ossification of the dermis and subcutaneous tissues. Diagnosis involves identifying the clinical features and genetic or epigenetic alterations affecting the GNAS locus. GNAS inactivation can be caused by pathogenic variants, imprinting issues, epimutations, or paternal 20q disomy. PPHP and POH/OC are caused by pathogenic variants of the paternal GNAS allele [1].
PHP-1b is a rare imprinting disorder characterized by renal parathyroid hormone (PTH) resistance, but the absence of physical features of AHO [2]. This distinguishes PHP-Ib from PHP-Ia, which results from mutations in the GNAS gene encoding the G protein α subunit. The autosomal-dominant form of PHP-Ib (ADPHP-Ib) is linked to chromosome 20q13.3, where the GNAS locus is located. Notably, loss of methylation of exon the A/B differentially methylated region (DMR) of GNAS has been observed in both sporadic PHP-Ib cases and affected members of AD-PHP-Ib families. Recent findings indicate that affected individuals from 12 unrelated AD-PHP-Ib families and four patients with sporadic PHP-Ib, but not healthy controls, are heterozygous for an approximately 3 kb microdeletion approximately 220 kb upstream of GNAS exon A/B. This deleted region includes three exons of the STX16 gene, which does not show any imprinting. Microdeletion leads to loss of methylation at exons A/B, without affecting other GNAS DMRs, suggesting that it disrupts the cis-acting element necessary for exon A/B methylation, causing renal PTH resistance in AD-PHP-Ib [3].
However, there has been no research on the causes of recurrent 3-kb microdeletion. Therefore, we aimed to elucidate the putative mechanism through genomic characterization of the 3-kb microdeletion.
Ten unrelated patients with PHP who visited Seoul National University Hospital between March 2009 and January 2019 were enrolled in the Korean Genetic Diagnosis Program for Rare Diseases Phase II [4]. This study was conducted in accordance with the tenets of the Declaration of Helsinki, and the protocol was approved by the Institutional Review Board of the Seoul National University Hospital. Informed consent was obtained from all enrolled participants.
To investigate the genetic and epigenetic changes in the samples, the MS-MLPA technique was employed using the SALSA MSMLPA probemix ME031-B2 GNAS kit, following the manufacturer’ s instructions (MRC-Holland, Amsterdam, Netherlands). Genomic DNA (200 ng) was denatured at 98°C for 5 minutes and hybridized with the ME031-B2 probe mix for 16 hours at 60°C. After hybridization, the product was divided into two tubes for copy number and methylation analyses using a methylation-sensitive endonuclease. The PCR products were analyzed on an ABI 3130xl capillary sequencer (Applied Biosystems, Waltham, MA, USA), and the data were processed using GeneMarker v.1.51 software (SoftGenetics, State College, PA, USA). Peak intensities were normalized to internal control probes, and the intensity ratios of identical probes from the samples were compared with those of the controls.
PCR primers were designed based on the consensus sequences from the split-read junctions. The amplified products were sequenced using an ABI PRISM 3730 xl DNA Analyzer (Applied Biosystems) with the BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems). Sequence analysis was conducted using Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA).
Repetitive elements within the breakpoint regions were identified using RepeatMasker in the UCSC Genome Browser (https://genome.ucsc.edu/). Sequence similarities were assessed using ClustalW (https://www.genome.jp/tools-bin/clustalw).
Table 1 summarizes the 10 cases, their classifications, and the genetic testing results. Table 1 comprises data for individuals classified as having PHP-1a and PHP-1b. Genetic testing revealed various abnormalities, such as paternal 20q disomy, STX16 2,979bp deletion, and mutations in the GNAS gene. Some patients exhibited no detectable genetic abnormalities.
A 2,979 bp deletion of STX16 was detected in two of the 10 probands (20%). Heterozygous deletions in exons 5 and 6 of the STX16 gene were initially identified using MLPA. The deletion of exon 7, which was not detected using MLPA, was suspected based on the results of an in-house copy number variation screening method [5]. Subsequently, junction PCR was performed. Finally, we confirmed a 2,979 bp deletion (chr20:57,243,566–57,246,545) encompassing exons 5–7 (Fig. 1).
When analyzing similarity using ClustalW within 1-kb upstream and downstream of the breakpoints, a 319 bp homologous region was detected, which showed only 2 bp mismatches, as depicted in Fig. 2 (bottom). This region contains one of the mammalian-wide interspersed repeats (MIRs), MIRb, which spans 191 bp. A 1-bp difference was observed between the upstream and downstream MIRb sequences. The upstream homologous sequence was located within intron 4 of STX16, whereas the downstream homologous sequence was located within intron 7.
STX16 gene deletion was detected in two of the 10 probands (20%). This deletion range is consistent with previous reports [6-10]. Most genomic rearrangements occur through mechanisms such as non-allelic homologous recombination (NAHR), nonhomologous end joining, fork stalling, and template switching [11]. However, the presence of homologous sequences in proximity to breakpoints favors rearrangements mediated by NAHR [12]. While NAHR mediated by Alu elements is well documented [13, 14], to the best of our knowledge, this is the first cases that NAHR via MIRs is a putative mechanism of rearrangement.
Short interspersed elements are widespread in mammalian genomes. The significant diversity of these repeats across placental orders suggests independent amplification of each lineage following mammalian radiation. Here, we introduce an ancient family of repeats, termed MIRs, whose sequence variation and widespread presence among placental mammals, marsupials, and monotremes indicate amplification during the Mesozoic era. With approximately 120,000 copies still identifiable in the human genome (0.2–0.3% of DNA), MIRs serve as a “fossilized” record of a substantial genetic event preceding the radiation of placental orders [15].
However, the evolutionary role of repeated element sequences in genomes remains debatable. A recent study suggested that an expanded repertoire of repeat elements may enhance organismal fitness by promoting somatic diversity, akin to the beneficial genetic diversity observed in microbial populations. Specifically, the characterization of somatic recombination involving Alu and L1 in this study provides a foundation for future investigations into the dynamics of somatic NAHR events and their implications for genome structure and function [16]. We believe that our study will contribute to broadening our understanding of the mechanisms underlying genomic rearrangements.
In conclusion, the findings of this study suggest that the common 2,979 deletion of STX16 is mediated by MIR-mediated nonallelic homologous recombination.
REFERENCES
1. Haldeman-Englert CR, Hurst ACE, Levine MA. Adam MP, Feldman J, Mirzaa GM, editors. 1993-2024. Disorders of GNAS Inactivation. 2017 Oct 26. GeneReviews® [Internet]. University of Washington, Seattle;Seattle (WA): Available from: https://www.ncbi.nlm.nih.gov/books/NBK459117/.
2. Mantovani G, Bastepe M, Monk D, de Sanctis L, Thiele S, Usardi A, et al. 2018; Diagnosis and management of pseudohypoparathyroidism and related disorders: first international Consensus Statement. Nat Rev Endocrinol. 14:476–500. DOI: 10.1038/s41574-018-0042-0. PMID: 29959430. PMCID: PMC6541219.
3. Bastepe M, Fröhlich LF, Hendy GN, Indridason OS, Josse RG, Koshiyama H, et al. 2003; Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 112:1255–63. DOI: 10.1172/JCI19159.
4. Kim MJ, Kim B, Lee H, Lee JS, Chae SW, Shin HS, et al. 2023; The Korean Genetic Diagnosis Program for Rare Disease Phase II: outcomes of a 6-year national project. Eur J Hum Genet. 31:1147–53. DOI: 10.1038/s41431-023-01415-8. PMID: 37414863. PMCID: PMC10545669.
5. Kim MJ, Lee S, Yun H, Cho SI, Kim B, Lee JS, et al. 2022; Consistent count region-copy number variation (CCR-CNV): an expandable and robust tool for clinical diagnosis of copy number variation at the exon level using next-generation sequencing data. Genet Med. 24:663–72. DOI: 10.1016/j.gim.2021.10.025. PMID: 34906491.
6. Chu X, Zhu Y, Wang O, Nie M, Quan T, Xue Y, et al. 2018; Clinical and genetic characteristics of pseudohypoparathyroidism in the Chinese population. Clin Endocrinol (Oxf). 88:285–94. DOI: 10.1111/cen.13516.
7. Danzig J, Li D, Jan de Beur S, Levine MA. 2021; High-throughput molecular analysis of pseudohypoparathyroidism 1b patients teveals novel genetic and epigenetic defects. J Clin Endocrinol Metab. 106:e4603–20. DOI: 10.1210/clinem/dgab460.
8. Elli FM, de Sanctis L, Peverelli E, Bordogna P, Pivetta B, Miolo G, et al. 2014; Autosomal dominant pseudohypoparathyroidism type Ib: a novel inherited deletion ablating STX16 causes loss of imprinting at the A/B DMR. J Clin Endocrinol Metab. 99:E724–8. DOI: 10.1210/jc.2013-3704. PMID: 24438374.
9. Kiuchi Z, Reyes M, Brickman AS, Jüppner H. 2021; A distinct variant of pseudohypoparathyroidism (PHP) first characterized some 41 years ago is caused by the 3-kb STX16 deletion. JBMR Plus. 5:e10505. DOI: 10.1002/jbm4.10505. PMID: 34258502. PMCID: PMC8260810. PMID: f5e78811d5224d85b7f9dad80d62fd49.
10. Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M. 2005; A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 76:804–14. DOI: 10.1086/429932. PMID: 15800843. PMCID: PMC1199370.
11. Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009; Mechanisms of change in gene copy number. Nat Rev Genet. 10:551–64. DOI: 10.1038/nrg2593.
12. Kataoka M, Aimi Y, Yanagisawa R, Ono M, Oka A, Fukuda K, et al. 2013; Alu-mediated nonallelic homologous and nonhomologous recombination in the BMPR2 gene in heritable pulmonary arterial hypertension. Genet Med. 15:941–7. DOI: 10.1038/gim.2013.41.
13. Ferlini A, Galié N, Merlini L, Sewry C, Branzi A, Muntoni F. 1998; A novel Alu-like element rearranged in the dystrophin gene causes a splicing mutation in a family with X-linked dilated cardiomyopathy. Am J Hum Genet. 63:436–46. DOI: 10.1086/301952.
14. Montagna M, Santacatterina M, Torri A, Menin C, Zullato D, Chieco-Bianchi L, et al. 1999; Identification of a 3 kb Alu-mediated BRCA1 gene rearrangement in two breast/ovarian cancer families. Oncogene. 18:4160–5. DOI: 10.1038/sj.onc.1202754.
15. Jurka J, Zietkiewicz E, Labuda D. 1995; Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 23:170–5. DOI: 10.1093/nar/23.1.170. PMID: 7870583. PMCID: PMC306646.
16. Pascarella G, Hon CC, Hashimoto K, Busch A, Luginbühl J, Parr C, et al. 2022; Recombination of repeat elements generates somatic complexity in human genomes. Cell. 185:3025–40. e6. DOI: 10.1016/j.cell.2022.06.032. PMID: 35882231.
Fig. 1
Detection of the 2,979 bp deletion of STX16. Heterozygous deletion of exons 5 and 6 of the STX16 gene was initially identified using multiplex ligation-dependent probe amplification (MLPA). The deletion of exon 7, which was not detected using MLPA, was suspected base on the results of an in-house method. Junction PCR was subsequently performed. Finally, we confirmed the 2,979 bp deletion encompassing exon 5 to exon 7.
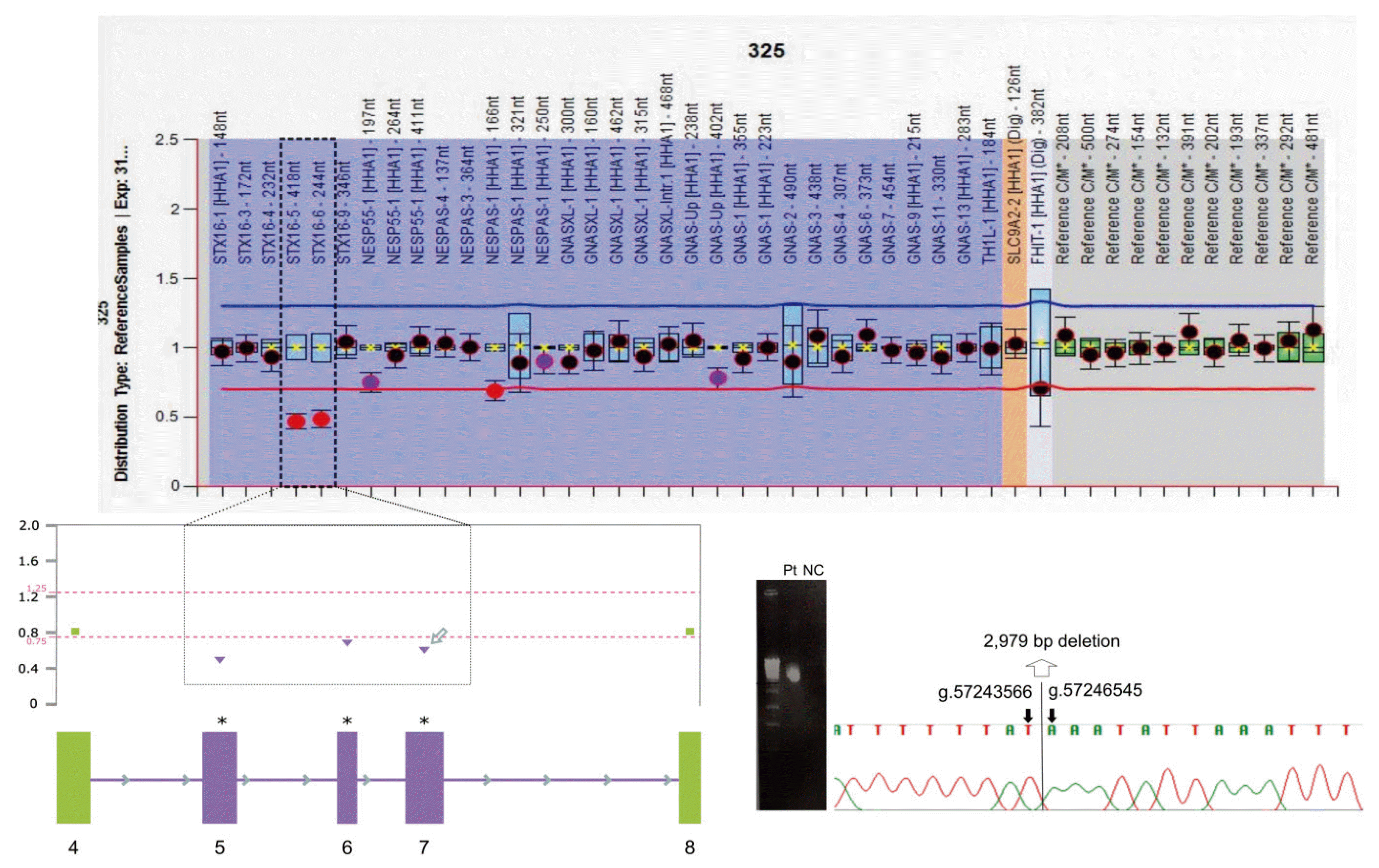
Fig. 2
Junction analysis of the 2,979 bp deletion of STX16. The distribution of repetitive elements around the 2,979 deletion of STX16 is shown. Two MIRbs are indicated by green squares. Below, the 319 bp homology sequence is displayed. The green text corresponds to MIRb, and the blue text represents the remaining sequences. The upstream homologous sequences and the downstream homologous sequences were completely identical except for 2 bp.
Abbreviation: MIRb, mammalian-wide interspersed repeat b.
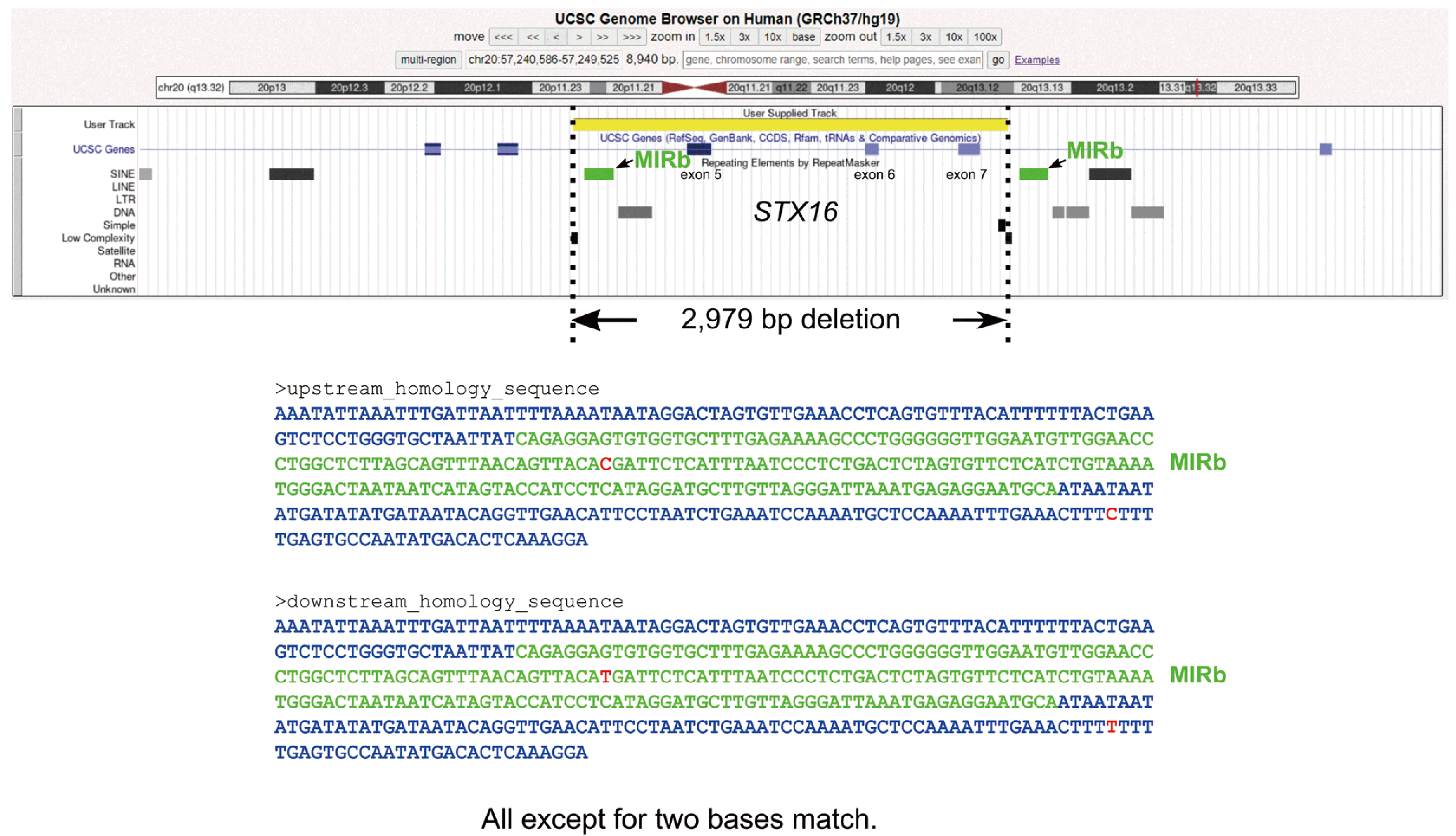
Table 1
Results of mutation analysis




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download