1. Liu Z, Davidson A. 2012; Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 18:871–82. DOI:
10.1038/nm.2752. PMID:
22674006. PMCID:
PMC3607103.
2. Barber MRW, Drenkard C, Falasinnu T, Hoi A, Mak A, Kow NY, et al. 2021; Global epidemiology of systemic lupus erythematosus. Nat Rev Rheumatol. 17:515–32. DOI:
10.1038/s41584-021-00668-1. PMID:
34345022. PMCID:
PMC8982275.
3. Izmirly PM, Wan I, Sahl S, Buyon JP, Belmont HM, Salmon JE, et al. 2017; The incidence and prevalence of systemic lupus erythematosus in New York County (Manhattan), New York: the Manhattan lupus surveillance program. Arthritis Rheumatol. 69:2006–17. DOI:
10.1002/art.40192. PMID:
28891252. PMCID:
PMC11102806.
4. Lewis MJ, Jawad AS. 2017; The effect of ethnicity and genetic ancestry on the epidemiology, clinical features and outcome of systemic lupus erythematosus. Rheumatology (Oxford). 56(Suppl 1):i67–77. DOI:
10.1093/rheumatology/kew399. PMID:
27940583.
5. Askanase A, Khalili L, Tang W, Mertz P, Scherlinger M, Sebbag E, et al. 2023; New and future therapies: changes in the therapeutic armamentarium for SLE. Best Pract Res Clin Rheumatol. 37:101865. DOI:
10.1016/j.berh.2023.101865. PMID:
37633826.
6. Han Y, Liu L, Zang B, Liang R, Zhao X, Liu B. 2023; Advances in natural products and antibody drugs for SLE: new therapeutic ideas. Front Pharmacol. 14:1235440. DOI:
10.3389/fphar.2023.1235440. PMID:
37492083. PMCID:
PMC10363611.
7. Appel GB, Contreras G, Dooley MA, Ginzler EM, Isenberg D, Jayne D, et al. Aspreva Lupus Management Study Group. 2009; Mycophenolate mofetil versus cyclophosphamide for induction treatment of lupus nephritis. J Am Soc Nephrol. 20:1103–12. DOI:
10.1681/ASN.2008101028. PMID:
19369404. PMCID:
PMC2678035.
8. Isenberg D, Appel GB, Contreras G, Dooley MA, Ginzler EM, Jayne D, et al. 2010; Influence of race/ethnicity on response to lupus nephritis treatment: the ALMS study. Rheumatology (Oxford). 49:128–40. DOI:
10.1093/rheumatology/kep346. PMID:
19933596. PMCID:
PMC2789586.
9. Merrill JT, Neuwelt CM, Wallace DJ, Shanahan JC, Latinis KM, Oates JC, et al. 2010; Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 62:222–33. DOI:
10.1002/art.27233. PMID:
20039413. PMCID:
PMC4548300.
10. Catalina MD, Bachali P, Yeo AE, Geraci NS, Petri MA, Grammer AC, et al. 2020; Patient ancestry significantly contributes to molecular heterogeneity of systemic lupus erythematosus. JCI Insight. 5:e140380. DOI:
10.1172/jci.insight.140380. PMID:
32759501. PMCID:
PMC7455079.
11. Wolpoff MH, Hawks J, Frayer DW, Hunley K. 2001; Modern human ancestry at the peripheries: a test of the replacement theory. Science. 291:293–7. DOI:
10.1126/science.291.5502.293. PMID:
11209077.
14. Hunter P. 2014; The genetics of human migrations: our ancestors migration out of Africa has left traces in our genomes that explain how they adapted to new environments. EMBO Rep. 15:1019–22. DOI:
10.15252/embr.201439469. PMID:
25216943. PMCID:
PMC4253842.
16. Karlsson EK, Kwiatkowski DP, Sabeti PC. 2014; Natural selection and infectious disease in human populations. Nat Rev Genet. 15:379–93. DOI:
10.1038/nrg3734. PMID:
24776769. PMCID:
PMC4912034.
17. Casanova JL, Abel L. 2005; Inborn errors of immunity to infection: the rule rather than the exception. J Exp Med. 202:197–201. DOI:
10.1084/jem.20050854. PMID:
16027233. PMCID:
PMC2212996.
18. Allison AC. 1954; Protection afforded by sickle-cell trait against subtertian malarial infection. Br Med J. 1:290–4. DOI:
10.1136/bmj.1.4857.290.
19. Parkes M, Cortes A, Van Heel DA, Brown MA. 2013; Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 14:661–73. DOI:
10.1038/nrg3502. PMID:
23917628.
20. Abel L, Alcaïs A, Schurr E. 2014; The dissection of complex susceptibility to infectious disease: bacterial, viral and parasitic infections. Curr Opin Immunol. 30:72–8. DOI:
10.1016/j.coi.2014.07.002. PMID:
25083600.
21. Quintana-Murci L, Clark AG. 2013; Population genetic tools for dissecting innate immunity in humans. Nat Rev Immunol. 13:280–93. DOI:
10.1038/nri3421. PMID:
23470320. PMCID:
PMC4015519.
22. Konstantinoudis G, Cameletti M, Gómez-Rubio V, Gómez IL, Pirani M, Baio G, et al. 2022; Regional excess mortality during the 2020 COVID-19 pandemic in five European countries. Nat Commun. 13:482. DOI:
10.1038/s41467-022-28157-3. PMID:
35079022. PMCID:
PMC8789777.
23. Landoni G, Maimeri N, Fedrizzi M, Fresilli S, Kuzovlev A, Likhvantsev V, et al. 2021; Why are Asian countries outperforming the Western world in controlling COVID-19 pandemic? Pathog Glob Health. 115:70–2. DOI:
10.1080/20477724.2020.1850982. PMID:
33241776. PMCID:
PMC7850376.
24. Fodil N, Langlais D, Gros P. 2016; Primary immunodeficiencies and inflammatory disease: a growing genetic intersection. Trends Immunol. 37:126–40. DOI:
10.1016/j.it.2015.12.006. PMID:
26791050. PMCID:
PMC4738049.
25. Langefeld CD, Ainsworth HC, Cunninghame Graham DS, Kelly JA, Comeau ME, Marion MC, et al. 2017; Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun. 8:16021. DOI:
10.1038/ncomms16021. PMID:
28714469. PMCID:
PMC5520018.
26. Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Riva S, Clerici M, et al. 2009; Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J Exp Med. 206:1395–408. DOI:
10.1084/jem.20082779. PMID:
19468064. PMCID:
PMC2715056.
27. Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, et al. 2010; Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 86:970–7. DOI:
10.1016/j.ajhg.2010.05.004. PMID:
20560212. PMCID:
PMC3032060.
28. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. 2012; Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 491:119–24. DOI:
10.1038/nature11582. PMID:
23128233. PMCID:
PMC3491803.
29. Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. 2005; Pathogen-driven selection and worldwide HLA class I diversity. Curr Biol. 15:1022–7. DOI:
10.1016/j.cub.2005.04.050. PMID:
15936272.
32. Chen H, Hayashi G, Lai OY, Dilthey A, Kuebler PJ, Wong TV, et al. 2012; Psoriasis patients are enriched for genetic variants that protect against HIV-1 disease. PLoS Genet. 8:e1002514. DOI:
10.1371/journal.pgen.1002514. PMID:
22577363. PMCID:
PMC3343879.
33. Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. 2017; The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 18:76. DOI:
10.1186/s13059-017-1207-1. PMID:
28449694. PMCID:
PMC5406920.
34. Ritari J, Koskela S, Hyvärinen K, FinnGen , Partanen J. 2022; HLA-disease association and pleiotropy landscape in over 235,000 Finns. Hum Immunol. 83:391–8. DOI:
10.1016/j.humimm.2022.02.003. PMID:
35221124.
35. Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. 2012; Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 44:1341–8. DOI:
10.1038/ng.2467. PMID:
23143594. PMCID:
PMC3510312.
36. Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. 2013; Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 45:730–8. DOI:
10.1038/ng.2667. PMID:
23749187. PMCID:
PMC3757343.
37. Diogo D, Bastarache L, Liao KP, Graham RR, Fulton RS, Greenberg JD, et al. 2015; TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PloS one. 10:e0122271. DOI:
10.1371/journal.pone.0122271. PMID:
25849893. PMCID:
PMC4388675.
38. Dendrou CA, Cortes A, Shipman L, Evans HG, Attfield KE, Jostins L, et al. 2016; Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med. 8:363ra149. DOI:
10.1126/scitranslmed.aag1974. PMID:
27807284. PMCID:
PMC5737835.
39. Boisson-Dupuis S, Ramirez-Alejo N, Li Z, Patin E, Rao G, Kerner G, et al. 2018; Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common
TYK2 missense variant. Sci Immunol. 3:eaau8714. DOI:
10.3410/f.734676937.793572033. PMID:
30578352. PMCID:
PMC6341984.
40. Kerner G, Ramirez-Alejo N, Seeleuthner Y, Yang R, Ogishi M, Cobat A, et al. 2019; Homozygosity for
TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc Natl Acad Sci U S A. 116:10430–4. DOI:
10.1073/pnas.1903561116. PMID:
31068474. PMCID:
PMC6534977.
41. Kerner G, Laval G, Patin E, Boisson-Dupuis S, Abel L, Casanova JL, et al. 2021; Human ancient DNA analyses reveal the high burden of tuberculosis in Europeans over the last 2,000 years. Am J Hum Genet. 108:517–24. DOI:
10.1016/j.ajhg.2021.02.009. PMID:
33667394. PMCID:
PMC8008489.
42. Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, et al. 2003; Human susceptibility and resistance to Norwalk virus infection. Nat Med. 9:548–53. DOI:
10.1038/nm860. PMID:
12692541.
43. Panda D, Gjinaj E, Bachu M, Squire E, Novatt H, Ozato K, et al. 2019; IRF1 maintains optimal constitutive expression of antiviral genes and regulates the early antiviral response. Front Immunol. 10:1019. DOI:
10.3389/fimmu.2019.01019. PMID:
31156620. PMCID:
PMC6529937.
44. Nunes-Santos CJ, Kuehn HS, Rosenzweig SD. 2020; IKAROS family zinc finger 1-associated diseases in primary immunodeficiency patients. Immunol Allergy Clin North Am. 40:461–70. DOI:
10.1016/j.iac.2020.04.004. PMID:
32654692. PMCID:
PMC7394939.
45. Rascovan N, Sjögren KG, Kristiansen K, Nielsen R, Willerslev E, Desnues C, et al. 2019; Emergence and spread of basal lineages of
Yersinia pestis during the Neolithic decline. Cell. 176:295–305.e10. DOI:
10.1016/j.cell.2018.11.005. PMID:
30528431.
46. Andrades Valtueña A, Neumann GU, Spyrou MA, Musralina L, Aron F, Beisenov A, et al. 2022; Stone Age
Yersinia pestis genomes shed light on the early evolution, diversity, and ecology of plague. Proc Natl Acad Sci U S A. 119:e2116722119. DOI:
10.3410/f.742004133.793592836. PMID:
35412864. PMCID:
PMC9169917.
48. Spurgin LG, Richardson DS. 2010; How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 277:979–88. DOI:
10.1098/rspb.2009.2084. PMID:
20071384. PMCID:
PMC2842774.
49. Satta Y, O'HUigin C, Takahata N, Klein J. 1994; Intensity of natural selection at the major histocompatibility complex loci. Proc Natl Acad Sci U S A. 91:7184–8. DOI:
10.1073/pnas.91.15.7184. PMID:
8041766. PMCID:
PMC44363.
50. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. 2015; The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43:D423–31. DOI:
10.1093/nar/gku1161. PMID:
25414341. PMCID:
PMC4383959.
54. Fernández-Viña MA, Klein JP, Haagenson M, Spellman SR, Anasetti C, Noreen H, et al. 2013; Multiple mismatches at the low expression HLA loci DP, DQ, and DRB3/4/5 associate with adverse outcomes in hematopoietic stem cell transplantation. Blood. 121:4603–10. DOI:
10.1182/blood-2013-02-481945. PMID:
23596045. PMCID:
PMC3668493.
55. Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. 2013; HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 191:5831–9. DOI:
10.4049/jimmunol.1302101. PMID:
24190657. PMCID:
PMC3872965.
56. Kaufman J. 2018; Generalists and specialists: a new view of how MHC class I molecules fight infectious pathogens. Trends Immunol. 39:367–79. DOI:
10.1016/j.it.2018.01.001. PMID:
29396014. PMCID:
PMC5929564.
57. Manczinger M, Boross G, Kemény L, Müller V, Lenz TL, Papp B, et al. 2019; Pathogen diversity drives the evolution of generalist MHC-II alleles in human populations. PLoS Biol. 17:e3000131. DOI:
10.1371/journal.pbio.3000131. PMID:
30703088. PMCID:
PMC6372212.
58. Crump JA, Luby SP, Mintz ED. 2004; The global burden of typhoid fever. Bull World Health Organ. 82:346–53. PMID:
15298225. PMCID:
PMC2622843.
59. Sun H, Yang Z, Lin K, Liu S, Huang K, Wang X, et al. 2015; The adaptive change of HLA-DRB1 allele frequencies caused by natural selection in a Mongolian population that migrated to the South of China. PLoS One. 10:e0134334. DOI:
10.1371/journal.pone.0134334. PMID:
26230582. PMCID:
PMC4521750.
60. Villesen P, Aagaard L, Wiuf C, Pedersen FS. 2004; Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 1:32. DOI:
10.1186/1742-4690-1-32. PMID:
15476554. PMCID:
PMC524368.
61. Belshaw R, Pereira V, Katzourakis A, Talbot G, Pačes J, Burt A, et al. 2004; Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A. 101:4894–9. DOI:
10.1073/pnas.0307800101. PMID:
15044706. PMCID:
PMC387345.
63. Stearrett N, Dawson T, Rahnavard A, Bachali P, Bendall ML, Zeng C, et al. 2021; Expression of human endogenous retroviruses in systemic lupus erythematosus: multiomic integration with gene expression. Front Immunol. 12:661437. DOI:
10.3389/fimmu.2021.661437. PMID:
33986751. PMCID:
PMC8112243.
64. Adelman MK, Marchalonis JJ. 2002; Endogenous retroviruses in systemic lupus erythematosus: candidate lupus viruses. Clin Immunol. 102:107–16. DOI:
10.1006/clim.2001.5153. PMID:
11846452.
66. Mustelin T, Ukadike KC. 2020; How retroviruses and retrotransposons in our genome may contribute to autoimmunity in rheumatological conditions. Front Immunol. 11:593891. DOI:
10.3389/fimmu.2020.593891. PMID:
33281822. PMCID:
PMC7691656.
68. Talotta R, Atzeni F, Laska MJ. 2020; Retroviruses in the pathogenesis of systemic lupus erythematosus: are they potential therapeutic targets? Autoimmunity. 53:177–91. DOI:
10.1080/08916934.2020.1755962. PMID:
32321325.
69. Perl A, Colombo E, Dai H, Agarwal R, Mark KA, Banki K, et al. 1995; Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum. 38:1660–71. DOI:
10.1002/art.1780381119. PMID:
7488288.
70. Chen CJ, Lin KH, Lin SC, Tsai WC, Yen JH, Chang SJ, et al. 2005; High prevalence of immunoglobulin A antibody against Epstein-Barr virus capsid antigen in adult patients with lupus with disease flare: case control studies. J Rheumatol. 32:44–7. PMID:
15630723.
71. Marchini J, Howie B. 2010; Genotype imputation for genome-wide association studies. Nat Rev Genet. 11:499–511. DOI:
10.1038/nrg2796. PMID:
20517342.
72. Guerra SG, Vyse TJ, Cunninghame Graham DS. 2012; The genetics of lupus: a functional perspective. Arthritis Res Ther. 14:211. DOI:
10.1186/ar3844. PMID:
22640752. PMCID:
PMC3446495.
73. Yin X, Kim K, Suetsugu H, Bang SY, Wen L, Koido M, et al. 2021; Meta-analysis of 208370 East Asians identifies 113 susceptibility loci for systemic lupus erythematosus. Ann Rheum Dis. 80:632–40. DOI:
10.1136/annrheumdis-2020-219209. PMID:
33272962. PMCID:
PMC8053352.
74. Morris DL, Taylor KE, Fernando MM, Nititham J, Alarcón-Riquelme ME, Barcellos LF, et al. 2012; Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am J Hum Genet. 91:778–93. DOI:
10.1016/j.ajhg.2012.08.026. PMID:
23084292. PMCID:
PMC3487133.
75. Kim K, Bang SY, Lee HS, Okada Y, Han B, Saw WY, et al. 2014; The HLA-DRβ1 amino acid positions 11-13-26 explain the majority of SLE-MHC associations. Nat Commun. 5:5902. DOI:
10.1038/ncomms6902. PMID:
25533202.
76. Bang SY, Choi JY, Park S, Choi J, Hong SJ, Lee HS, et al. 2016; Brief report: influence of HLA-DRB1 susceptibility alleles on the clinical subphenotypes of systemic lupus erythematosus in Koreans. Arthritis Rheumatol. 68:1190–6. DOI:
10.1002/art.39539. PMID:
26663868.
77. Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. 2016; High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet. 48:323–30. DOI:
10.1038/ng.3496. PMID:
26808113. PMCID:
PMC4767573.
78. Gonzalez-Galarza FF, McCabe A, Santos EJMD, Jones J, Takeshita L, Ortega-Rivera ND, et al. 2020; Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 48:D783–8. DOI:
10.1093/nar/gkz1029. PMID:
31722398. PMCID:
PMC7145554.
79. Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, et al. 2016; Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet. 48:940–6. DOI:
10.1038/ng.3603. PMID:
27399966. PMCID:
PMC4966635.
80. Lessard CJ, Sajuthi S, Zhao J, Kim K, Ice JA, Li H, et al. 2016; Identification of a systemic lupus erythematosus risk locus spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol. 68:1197–209. DOI:
10.1002/art.39548. PMID:
26663301. PMCID:
PMC4981330.
81. Molineros JE, Looger LL, Kim K, Okada Y, Terao C, Sun C, et al. 2019; Amino acid signatures of HLA class-I and II molecules are strongly associated with SLE susceptibility and autoantibody production in Eastern Asians. PLoS Genet. 15:e1008092. DOI:
10.1371/journal.pgen.1008092. PMID:
31022184. PMCID:
PMC6504188.
83. Wallace DJ, Furie RA, Tanaka Y, Kalunian KC, Mosca M, Petri MA, et al. 2018; Baricitinib for systemic lupus erythematosus: a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet. 392:222–31. DOI:
10.1016/S0140-6736(18)31363-1. PMID:
30043749.
84. He J, Zhang R, Shao M, Zhao X, Miao M, Chen J, et al. 2020; Efficacy and safety of low-dose IL-2 in the treatment of systemic lupus erythematosus: a randomised, double-blind, placebo-controlled trial. Ann Rheum Dis. 79:141–9. DOI:
10.1136/annrheumdis-2019-215396. PMID:
31537547. PMCID:
PMC6937406.
85. Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, et al. 2020; Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med. 382:211–21. DOI:
10.1056/NEJMoa1912196. PMID:
31851795.
86. van Vollenhoven RF, Hahn BH, Tsokos GC, Lipsky P, Fei K, Gordon RM, et al. 2020; Maintenance of efficacy and safety of ustekinumab through one year in a phase II multicenter, prospective, randomized, double-blind, placebo-controlled crossover trial of patients with active systemic lupus erythematosus. Arthritis Rheumatol. 72:761–8. DOI:
10.1002/art.41179. PMID:
31769212.
87. Isenberg D, Furie R, Jones NS, Guibord P, Galanter J, Lee C, et al. 2021; Efficacy, safety, and pharmacodynamic effects of the Bruton's tyrosine kinase inhibitor fenebrutinib (GDC-0853) in systemic lupus erythematosus: results of a phase II, randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol. 73:1835–46. DOI:
10.1002/art.41811. PMID:
34042314.
88. Furie RA, Aroca G, Cascino MD, Garg JP, Rovin BH, Alvarez A, et al. 2022; B-cell depletion with obinutuzumab for the treatment of proliferative lupus nephritis: a randomised, double-blind, placebo-controlled trial. Ann Rheum Dis. 81:100–7. DOI:
10.1136/annrheumdis-2021-220920. PMID:
34615636. PMCID:
PMC8762029.
89. Morand E, Pike M, Merrill JT, van Vollenhoven R, Werth VP, Hobar C, et al. 2023; Deucravacitinib, a tyrosine kinase 2 inhibitor, in systemic lupus erythematosus: a phase II, randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol. 75:242–52. DOI:
10.1002/art.42391. PMID:
36369798. PMCID:
PMC10100399.
90. Webb R, Kelly JA, Somers EC, Hughes T, Kaufman KM, Sanchez E, et al. 2011; Early disease onset is predicted by a higher genetic risk for lupus and is associated with a more severe phenotype in lupus patients. Ann Rheum Dis. 70:151–6. DOI:
10.1136/ard.2010.141697. PMID:
20881011. PMCID:
PMC3034281.
91. Joo YB, Lim J, Tsao BP, Nath SK, Kim K, Bae SC. 2018; Genetic variants in systemic lupus erythematosus susceptibility loci, XKR6 and GLT1D1 are associated with childhood-onset SLE in a Korean cohort. Sci Rep. 8:9962. DOI:
10.1038/s41598-018-28128-z. PMID:
29967481. PMCID:
PMC6028392.
92. Reid S, Alexsson A, Frodlund M, Morris D, Sandling JK, Bolin K, et al. 2020; High genetic risk score is associated with early disease onset, damage accrual and decreased survival in systemic lupus erythematosus. Ann Rheum Dis. 79:363–9. DOI:
10.1136/annrheumdis-2019-216227. PMID:
31826855. PMCID:
PMC7034364.
93. Kwon YC, Ha E, Kwon HH, Park DJ, Shin JM, Joo YB, et al. 2023; Higher genetic risk loads confer more diverse manifestations and higher risk of lupus nephritis in systemic lupus erythematosus. Arthritis Rheumatol. 75:1566–72. DOI:
10.1002/art.42516. PMID:
37011055.
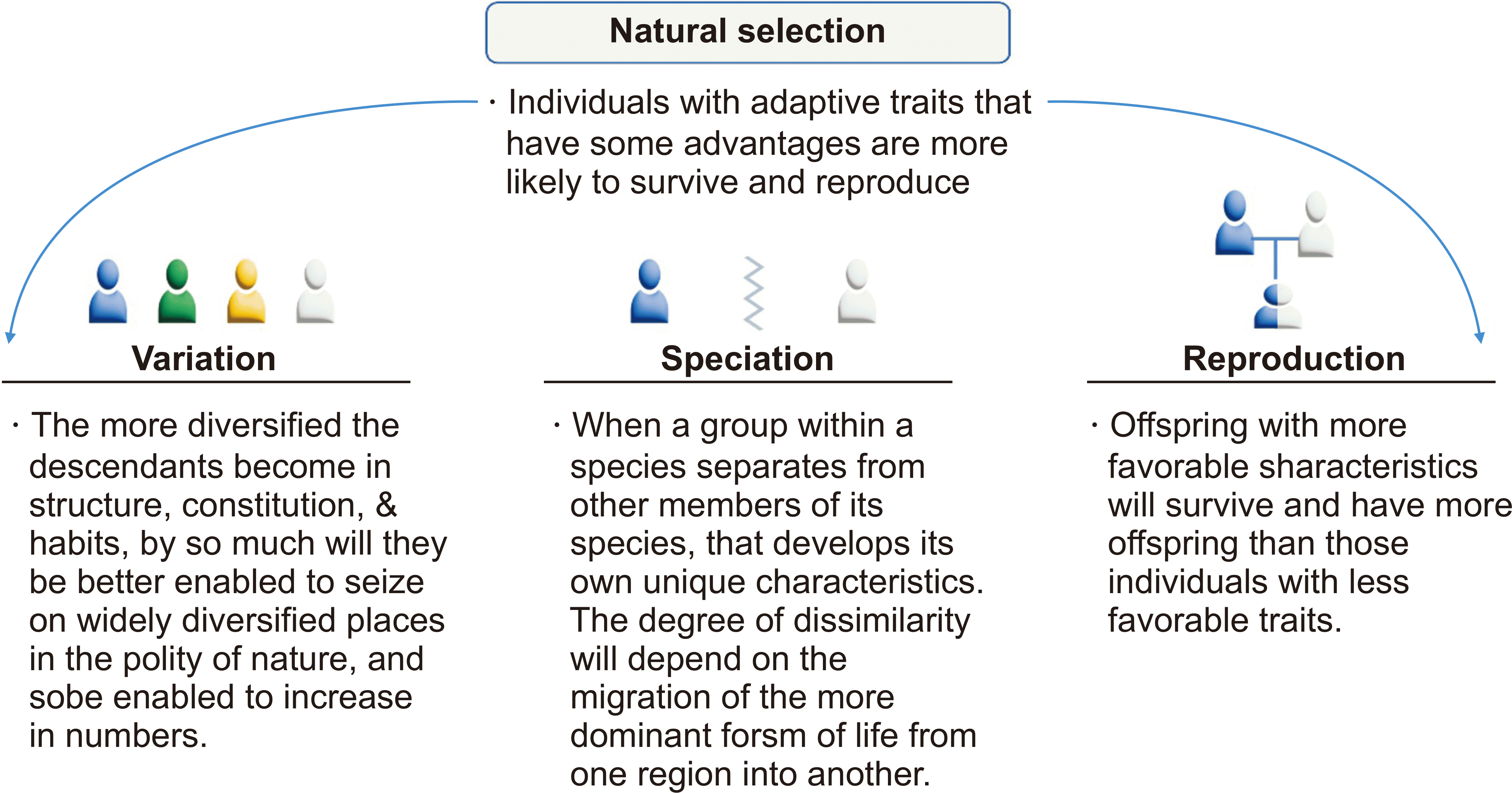
94. Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favored races in the struggle for life. John Murray;London: DOI:
10.5962/bhl.title.68064.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download