INTRODUCTION
MATERIALS AND METHODS
Eligibility criteria
Search strategy
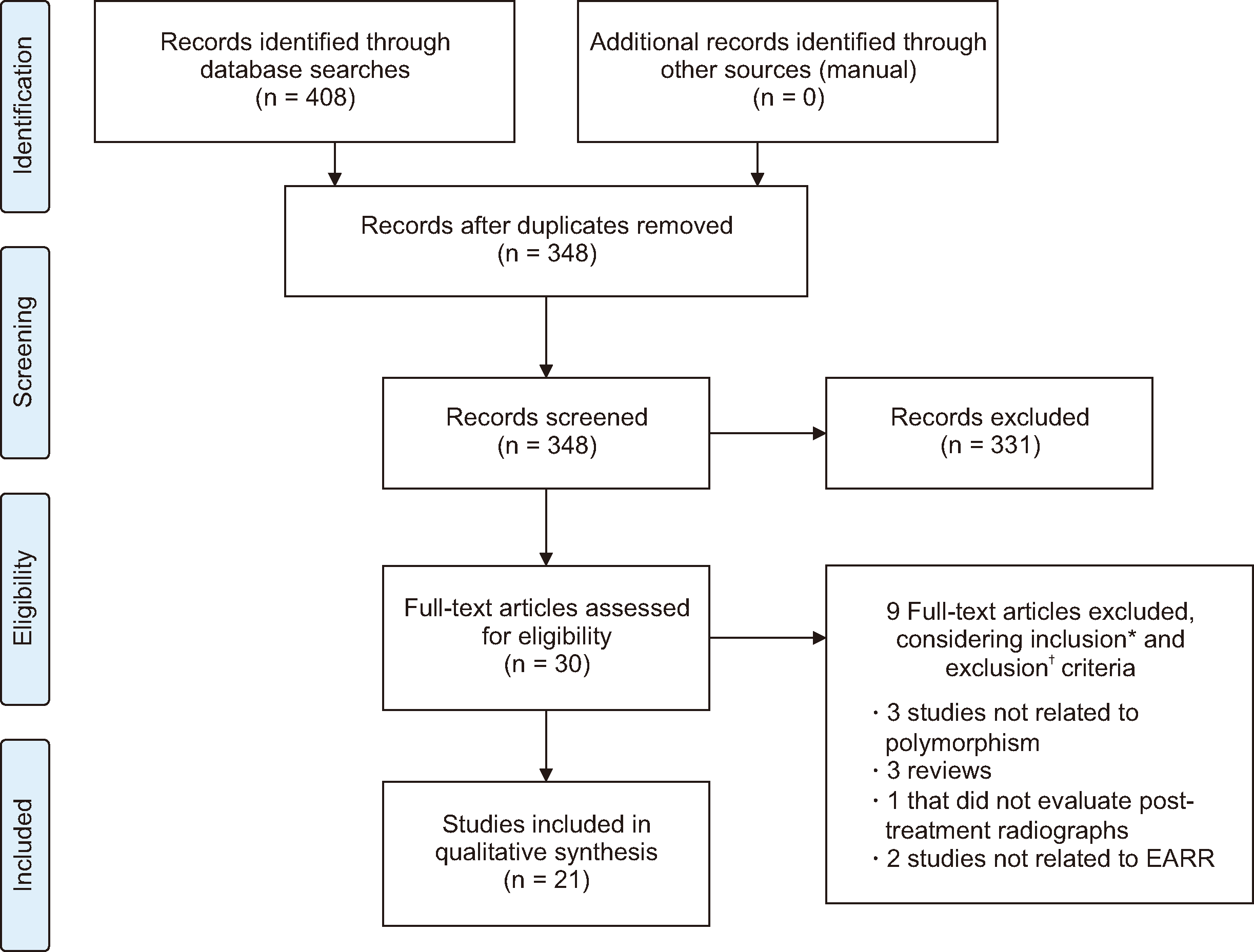 | Figure 1PRISMA flow diagram showing the process of article selection.
EARR, external apical root resorption.
*Inclusion criteria: epidemiological studies (cross-sectional, case-control, cohort and clinical trials) that evaluated the association of genetic polymorphisms and EARR in patients treated with fixed appliances; studies that used polymerase chain reaction-restriction fragment length polymorphism analysis to evaluate genetic polymorphisms; and, studies that reported pre- and post-orthodontic treatment.
†Exclusion criteria: reviews, articles or other systematic reviews; case reports; case series; descriptive studies; opinion articles; abstracts; letters to the editor; laboratory and/or animal studies; subjects with cleft lip, palate or both; studies on individuals with other craniofacial deformities/syndromes; studies on unrelated topics that had no association with genetics or those that presented data of EARR not caused by orthodontic treatment.
|
Data extraction
Table 1
| Study/country | Design | Population | Sample size analyzed (case) | Sample size analyzed (control) | Participant age (yr) | Tooth evaluation | Measurement criteria | Gene polymorphism | Result | Conclusion |
|---|---|---|---|---|---|---|---|---|---|---|
| Al-Qawasmi et al.1 (2003)/USA | Cross-sectional | University clinic and private orthodontic practice | 118 | - | 12.1 ± 1.89 | MxCI with the longest root, MndCI with the longest root, and the mesial and distal roots of both the MndFM | Ceph and PAN |
IL1A (rs1800587)
IL1B (rs1143634)
|
IL1β allele 1 had a 5.6-fold (95% CI: 1.9–21.2) increased risk of EARR greater than 2 mm compared with those who are not homozygous for the IL-1B allele 1 (P = 0.004) | IL1B polymorphisms are associated with EARR |
| Al-Qawasmi et al.9 (2003)/USA | Cross-sectional | University clinic and private orthodontic practice | 124 | - | 12.3 ± 1.82 | MxCI with the longest root, MndCI with the longest root, and the mesial and distal roots of both the MndFM | Ceph and PAN |
TNSALP (rs not informed)
TNFα (rs1800629)
TNFRSF11A (rs1805034)
|
TNFRSF11A (LOD = 2.5; P = 0.02) |
No evidence of linkage was found with EARR and the TNFα and TNSALP genes
TNFRSF11A is associated with EARR
|
| Bastos Lages et al.11 (2009)/Brazil | Case-control | Private orthodontic practice | 23 | 38 | 18.9 ± 5.2 | MxCI, MxLI | Periapical radio-graphs | IL1B (rs1143634) | 2/2 vs. 1/2 + 1/1, OR = 4.00; 95% CI:1.23–12.9; P = 0.0349 | The IL1B polymorphism associated with EARR |
| Tomoyasu et al.15 (2009)/Japan | Case-control | University clinic | 54 | - |
Male 19
Female 21
|
MxCI, MndCI, FM | Ceph and PAN | IL1B (rs1143634) |
IL1B
Maxillary central incisor (P = 0.47) Mandibular central incisor (P = 0.48) Mandibular first molar, metal root (P = 0.08) Mandibular first molar, distal root (P = 0.22)
|
No association |
| Iglesias-Linares et al.17 (2012)/Spain | Case-control | University clinic | 73 | 73 | 23.78 ± 5.91 | Maxillary root-filled PM and the contralateral tooth with a vital pulp | Ceph and PAN |
IL1B (rs1143634)
IL1A (rs1800587)
|
CC vs. CT/TT 5.143 1.38–19.10 (P = 1.00)
TT vs. CT/CC 2.032 1.99–14.773 (P = 0.031)
CT vs. CC/TT 10.66 0.72–158.50 (P = 0.625)
|
IL1B polymorphisms are associated with a twofold increased predisposition to have EARR secondary to orthodontic treatment in endodontically treated teeth |
| Linhartovaet al. 8 (2013)/Czech Republic | Case-control | University clinic | 32 | 74 |
15.0 ± 4.1
and
15.2 ± 5.3
|
MxCI, MxLI | Ceph and PAN |
IL1A (rs1800587)
IL1B (rs1143634)
IL1RN (86pb VNTR)
|
IL1RN
Variants in girls – short allele P = 0.020, OR = 2.50; 95% CI: 1.13–5.53
|
No significant role of IL1A and IL1B variants in EARR
IL1RN may be associated with EARR, especially in girls
|
| Iglesias-Linares et al.14 (2012)/Spain | Case-control | University clinic | 25 | 29 |
22.25 ± 5.30
and
23.89 ± 5.03
|
MxCI, MxLI with the longest root | Ceph and PAN |
IL1A (rs1800587)
IL1B (rs1143634)
IL1RN (rs419598)
|
IL1B
CC vs. CT⁄ TT OR = 3.477 (1.12–10.72); P = 0.027
IL1A
CC vs. CT⁄ TT OR = 2.51 (0.8–7.57); P = 0.097
IL1R
TT vs. CC⁄TC OR = 6.750 (2.04–22.27); P = 0.001
|
Variations in the IL1RN and not only in the IL1B gene are determinants of a predisposition to post-orthodontic EARR |
| Iglesias-Linares et al.16 (2012)/Spain | Case-control | University clinic | 39 | 54 |
24.54 ± 5.85
and
23.89 ± 5.72
|
Upper second root-filled PM | Ceph and PAN |
IL1A (rs1800587)
IL1B (rs1143634)
|
IL1B
CC vs. CT/TT OR: 2.54 (1.05–6.12); P = 0.035
TT vs. CT/CCOR: 11.59 (1.36–98.61); P = 0.006
CT vs. CC/TT OR: 0.12 (0.03–0.39); P = 0.001
|
IL1B gene polymorphisms are associated with EARR |
| Iglesias-Linares et al.22(2013)/Spain | Case-control | University clinic | 39 | 54 |
24.54 ± 5.85
and
23.89 ± 5.72
|
Upper second root-filled PM | Ceph and PAN | IL1RN (rs419598) |
IL1RN
CC vs. CT/TT OR: 10.857 (3.97–29.6); P = 0.001
CT vs. TT/CC OR: 0.18 (0.06–0.50); P = 0.001
CC vs. CT/CC OR: 0.10 (0.13–0.83); P = 0.011
|
IL1RN polymorphisms are associated with an increased risk of suffering post-orthodontic EARR in root-filled teeth |
| Iglesias-Linares et al.26 (2014)/Spain | Case-control | University clinic | 37 | 50 |
24.70 ± 5.95
and
23.80 ± 5.33
|
MxCI, MxLI with the longest root | Ceph and PAN | SPP1(rs9138, rs11730582) |
rs9138
CC vs. CA/AA OR: 4.10 1.03–16.35 (P = 0.045)
AA vs. CA/CC OR: 0.200.05–0.81 (P = 0.025)
CA vs. CC/AA OR: 0.8 0.20–3.53 (P = 0.823)
rs11730582
CC vs. CT/TTOR: 11.681.12–121.06 (P = 0.039)
TT vs. CT/CCOR: 0.450.07–2.80 (P = 0.39)
CT vs. CC/TTOR: 0.0350.062–0.90 (P = 0.035)
|
Specific allele (not informed) of the SPP1 is associated with genetic susceptibility to EARR |
| Sharab et al.27 (2015)/USA | Case-control | Private orthodontic practice | 67 | 67 |
15.78 ± 1.13
and
15.79 ± 1.14
|
MxCI, MxLI | Occlusal |
P2RX7 (rs208294, rs1718119, rs2230912)
CASP1/ICE (rs580253,rs530537)
CASP5 (rs554344)
IL1B (rs1143634)
IL1A (rs1800587)
IL1Ra (rs419598)
|
P2RX7, rs208294
CC: 28 (41.8%)
CT: 33 (49.3%)
TT: 6 (9.0%)
P = 0.0028
P2RX7, rs1718119
GG: 32 (47.8%)
GA: 25 (37.3%)
AA: 10 (14.9%)
P2RX7, rs2230912
AA: 53 (79.1%)
AG: 13 (19.4%)
GG: 1 (1.5%)
CASP1/ICE, rs530537
TT: 24 (35.8%)
TC: 30 (44.8%)
CC: 13 (19.4%)
CASP1/ICE, rs580253
GG: 52 (77.6%)
GA: 14 (20.9%)
AA: 1 (1.5%)
CASP5, rs554344
GG: 52 (77.6%)
GC: 14 (20.9%)
CC: 1 (1.5%)
IL1B, rs1143634
GG: 37 (55.2%)
GA: 26 (38.8%)
AA: 4 (6.0%)
IL1Ra, rs419598
TT: 41 (61.2%)
TC: 23 (34.3%)
CC: 3 (4.5%)
P = 0.0533
IL1A, rs1800587
GG: 28 (41.8%)
GA: 28 (41.8%)
AA: 11 (16.4%)
|
P2RX7 rs208294 was associated with EARR |
| Borilova Linhartova et al.28 (2017)/Czech Republic | Case-control | University clinic | 30 | 69 | 15.0 ± 4.7 | MxCI, MxLI rooth and crown lengths | Ceph and PAN | IL-17(rs2275913); SPP1 (rs11730582, rs9138); P2RX7(rs208294, Tyr155His, rs1718119); 11B (rs3102735, rs2073618) | No significant differences were observed in allele or genotype frequencies of all seven studied SNPs. Specific haplotype of P2RX7 (rs208294 and rs1718119) modified the risk of EARR development (P < 0.05) | The variability in the P2RX7 gene may be important factor contributing to the etiopatho-genesis of post-orthodontic EARR |
| Ciurla et al.25 (2021)/Poland | Case-control | Private orthodontic practice | 40 | 61 | 22.9 ± 6.3 | MxCI, MxLI, MndCI, MndLI, FM | Ceph and PAN | IL1RN (rs419598) and P2RX7 (rs208294) | P2RX7 (rs208294) and IL1RN (rs419598) modified the risk of EARR development (P < 0.05) | The analysis of the P2RX7 and IL1RN gene polymorphisms showed that the presence of SNPs of these genes may predispose individuals to EARR |
| Guo et al.23 (2016)/China | Case-control | University clinic | 174 | - | 14.07 ± 3.10 | MxCI | CBCT | IL1RN (rs419598); IL-6 SNP (rs1800796) | The IL-6 SNP rs1800796 GC was associated with EARR, and root resorption | IL-6 SNP rs1800796 GC is a risk factor for EARR |
| Ciurla et al.18 (2021)/Poland | Case-control | Private orthodontic practice | 101 | - | 21.32 ± 7.28 | MxCI, MxLI, MndCI, MndLI, FM | Ceph and PAN | IL-1β, TN- FRSF11B, CASP1, and IL-6 | A significant association was found between EARR presence and the SNP for the IL-1β gene but not for the TNFRSF11B, CASP1, and the IL-6 genes. IL-1β gene increases the odds of developing EARR by around four times | A significant association between EARR occurrence and the SNP for the IL-1β gene. Conversely, the effect of SNPs for CASP1, TNFRSF11B, and IL-6 genes on EARR presence was not confirmed by the present study |
| Borges de Castilhos et al.24 (2019)/Brazil | Case-control | University clinic and private orthodontic practice | 178 | 160 | 14.9 (8–21) | MxCI | Periapical radio-graphs | RANKL, RANK, OPG | For polymor-phisms of RANKL, no significant association was found with EARR. For RANK polymorphisms, only rs12455775 was associated with EARR Regarding OPG polymorphisms, an association of rs3102724, rs2875845, rs1032128, and rs3102728 with EARR was found | Regarding the analysis of polymorphisms in the genes RANKL, RANK, and OPG, several SNPs in RANK and OPG were associated with EARR, but only the association of the allele A of the rs3102724 in OPG remained after multivariate analysis |
| Silva et al.19 (2022)/Portugal | Cross-sectional | University clinic and private orthodontic practice | 195 | - |
< 14 (n = 63); 14–18
(n = 85);
18 > (n = 47)
|
MxCI, MxLI, MxCanine | PAN | rs1143634 (in IL1B gene) and rs3102735 (TNFRSF11B gene, encoding OPG) and (rs315952 from IL1RN), rs1805034 from TNFRSF11A, encoding RANK, and rs1718119 from P2RX7 | For genes encoding OPG, RANK and the IL1 and IL1RN, the effect of analyzed variants changed from protective to deleterious depending on the duration of treatment and the age of the patient | This work shows that in OIEARR the impact of genetic susceptibility factors is dynamic changing according to clinical variables |
| Iber-Díaz et al.29 (2020)/Spain | Cohort | University clinic | 101 | 361 |
21.52 ±
11.65
and
22.83 ± 11.66
|
MxCI, MxLI | PAN | Genes located in the X chromosome, specifically, STAG2 (rs151184635) and RP1-30E17.2 (rs55839915) |
Two novel putative genes located in the X chromosome, specifically, STAG2 gene, rs151184635 and RP1-30E17.2 gene, rs55839915, were associated with aggressive EARR
These variants were found to be associated with an increased risk of aEARR, particularly restricted to male. Marginal associations were found at previously studied variants such as SPP1: rs11730582, P2RX7: rs1718119, and TNFRSF11A: rs8086340, found solely in females
|
Multiple putative genetic variants located at chromosomes X and Y are potentially implicated in an extreme phenotype of aggressive EARR |
| Marañón-Vásquez et al.30 (2023)/Germany | Cross-sectional | University clinic and private orthodontic practice | 143 | - | 13.5 ± 4.5 | MxCI, FM | Ceph and PAN | VDR rs731236 – TaqI (A/G); VDR rs7975232 – ApaI (C/A); VDR rs1544410 – BsmI (T/C); VDR rs2228570 – FokI (A/G); GC rs4588 (T/G); CYP27B1 rs4646536 (G/A); CYP24A1 rs927650 (T/C) | Individuals carrying the AA genotype in VDR rs2228570 had a 21% higher EARRmol than those having AG and GG genotypes. EARRmol in heterozygous rs2228570, was 12% lower than for homozygotes. Participants with the CCG haplotype (rs1544410-rs7975232-rs731236) in VDR had an EARRmol 16% lower than those who did not carry this haplotype. Regarding CYP27B1 rs4646536, EARRinc in participants who had at least one G allele was 42% lower than for homozygotes AA | Although these results did not remain significant after multiple testing adjustment, potential associ- ations may still be suggested |
| Pereira et al.20 (2016)/Portugal | Cross-sectional | University clinic and private orthodontic practice | 195 | - | 17.24 ± 6.8 | MxCI, MxLI, MxCanine | PAN | IL1B (rs1143634), IL1RN (rs315952) IRAK1 (rs1059703) | Homozygosity/ hemizygosity for variant C from IRAK1 gene (P = 0.018) proved to be a protective factor | IRAK1 polymorphism is proposed as a protective variant for EARR |
| Iglesias-Linares et al.21 (2017)/Spain | Case-control | University clinic and private orthodontic practice | 174 | 198 |
28.48 ±
13.6
and
26.29 ± 13.66
|
MxCI, MxLI root | PAN | IL1B (rs1143634), IL1RN (rs419598), SPP1 (rs9138/rs11730582) | Only subjects homozygous for the T allele of IL1RN (rs419598) were more prone to OIEARR during orthodontic treatment | Only subjects homozygous for the T allele of IL1RN (rs419598) were more prone to OIEARR during orthodontic treatment |
MxCI, maxillary central incisor; MxLI, maxillary lateral incisor; MndCI, mandibular central incisor; MndFM, mandibular first molar; FM, first molar; PM, premolars; Ceph, lateral cephalogram; PAN, panoramic radiograph; EARRmol, external apical root resorption of the lower first molars; EARRinc, external apical root resorption of the upper central incisors; OR, odds ratio; CI, confidence interval; EARR, external apical root resorption; LOD, logarithm of odds; OIEARR, orthodontically induced external apical root resorption; VNTR, variable number tandem repeat; MndLI, mandibular central incisor; SNP, single-nucleotide polymorphism; CBCT, cone beam computed tomography; MxCanine, maxillary canine adenine.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download