Abstract
Few studies have focused on the association between clonal hematopoiesis of indeterminate potential (CHIP) and β-amyloid (Aβ) deposition in the brain, which causes Alzheimer’s disease. We aimed to investigate the potential role of CHIP in brain Aβ deposition in Korean patients. We enrolled 58 Korean patients over 50 yrs of age with cognitive impairment who underwent brain Aβ positron emission tomography. We explored CHIP in their peripheral blood using deep-targeted next-generation sequencing. Irrespective of the presence or absence of brain Aβ deposition, mutations in DNMT3A and the C:G>T:A single-nucleotide variants were identified as the primary characteristics, which reflect aged hematopoiesis in the study population. Multivariate logistic regression revealed that the presence of CHIP was not associated with brain Aβ deposition. As both CHIP and brain Aβ deposition are associated with aging, further research is required to elucidate their possible interplay.
Clonal hematopoiesis of indeterminate potential (CHIP) is characterized by the presence of somatic mutations in genes associated with hematologic malignancies in hematopoietic stem and progenitor cells (HSPCs) with a variant allele frequency (VAF) of ≥2% in the absence of overt hematologic malignancy or cytopenia [1]. The prevalence of these mutations, predominantly in DNMT3A, TET2, and ASXL1, increases with age [2]. Age-related clonal hematopoiesis is associated with an increased risk of hematologic cancer and cardiovascular disease, including coronary artery disease and ischemic stroke [2, 3]. In addition, recent studies have identified associations between CHIP and other diseases, including aortic valve stenosis [4, 5], chronic ischemic heart failure [6], chronic obstructive pulmonary disease [7, 8], and rheumatoid arthritis [9]. Notably, Bouzid, et al. [10] observed that individuals with CHIP had a reduced risk of developing Alzheimer’s disease (AD) over time, which also attenuated the effect of the APOE ε4 allele. Furthermore, they found a negative association between CHIP and AD neuropathological characteristics, including brain β-amyloid (Aβ) deposition and tau neurofibrillary tangles, as detected in autopsies.
In Korea, brain Aβ deposition in patients with cognitive impairment is often examined using amyloid positron emission tomography (PET) to evaluate AD or other causes of cognitive impairment. A positive amyloid PET scan does not necessarily confirm AD but indicates the presence of significant levels of Aβ, the hallmark pathology of AD, and suggests that AD is likely the cause of cognitive impairment in the patient [11-13]. Conversely, a negative amyloid PET reduces the likelihood that cognitive impairment is due to AD [11-13]. The previous study demonstrated the direct association between AD and CHIP; however, no study has investigated the association between these two factors using amyloid PET results. Therefore, we aimed to investigate the association between CHIP and brain Aβ deposition in the Korean population.
Fifty-eight patients with cognitive impairment were enrolled. All patients underwent 18F-labeled florbetaben amyloid brain PET-computed tomography (CT) using a Gemini TF 16 scanner (Philips Medical Systems, Cleveland, OH, USA), which was visually assessed based on regional cortical tracer uptake and brain amyloid plaque load (BAPL) scores [14]. BAPL scores of 2 and 3 indicate Aβ-positivity, whereas a score of 1 indicates Aβ-negativity. Ultimately, 43 patients who tested Aβ-positive and 15 who tested Aβ-negative were included in the study. Subjective cognitive decline (SCD), mild cognitive impairment (MCI), and dementia (i.e., major cognitive impairment) were diagnosed based on the criteria of the Subjective Cognitive Decline Initiative Working Group [15], Petersen’s criteria [16], and the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) [17], respectively. Age, cognitive status, and brain Aβ deposition were determined at the time of blood draw. All patients provided written informed consent. The study was reviewed and approved by the Institutional Review Board (IRB) of Chung-Ang University College of Medicine. Seoul, Korea (IRB No. 2207-012-514).
Peripheral blood DNA was used for APOE genotyping and next-generation sequencing (NGS). APOE genotyping was performed using real-time PCR analysis. PCRs were run using a Real-Q ApoE genotyping kit (BioSewoom, Seoul, Korea) on an ABI 7500 Real-time PCR System (Applied Biosystems, Waltham, MA, USA). CHIP was detected by deep-targeted NGS with a mean depth of 1,576 on an Ion Torrent system (Thermo Fisher, Waltham, MA, USA) using a panel of 61 hematological malignancy-related genes (Supplemental Data Table S1). As CHIP variants, single-nucleotide variants (SNVs) and small insertion-deletion variants with a VAF of ≥2% were filtered using an in-house filtering strategy (detailed in Supplemental Methods).
Student’s t-test and the Mann–Whitney test were used to compare continuous variables between groups. The chi-squared test, chi-squared test for trend, and Fisher’s exact test were used to compare categorical variables between groups. Multivariate logistic regression analysis was performed to evaluate the risk of brain Aβ deposition, and odds ratios (ORs) were calculated with 95% confidence intervals (CIs). Statistical analyses were conducted using R version 4.2.3 (The R Foundation for Statistical Computing, Vienna, Austria) and GraphPad Prism version 9.3.1 (GraphPad, San Diego, CA, USA). P<0.05 was considered statistically significant.
The demographics of the study population are presented in Table 1. The Aβ-positive group (N=43) tended to be more severely cognitively impaired (chi-squared test for trend, P=0.011) and comprised more APOE ε4 carriers (60.5% vs. 20.0%, P= 0.015) than the Aβ-negative group (N=15).
Of the 58 patients, nine patients (15.6%) harbored 11 CHIP variants in CBL, DNMT3A, JAK2, KMT2A, PRPF8, and TET2 (Table 2). As all patients included in this study showed complete blood counts within reference ranges, the possibility of undiagnosed blood cancer was ruled out. Seven patients (16.3%) were Aβ-positive, and two patients (13.2%) were Aβ-negative. The prevalence of CHIP in this study was similar to that in previous studies detecting CHIP in Korean individuals based on a VAF cut-off of 2% (6.5%–21%) [4, 18]. However, a previous study that utilized a VAF cut-off of 1% reported a higher prevalence of CHIP among Korean individuals (41%) [19]. The differences in CHIP prevalence may be attributed to not only the variant filtering strategy used but also heterogeneity in sample size, the sequencing method used, and the genes analyzed. Among the 11 CHIP variants, seven are reported in the Catalogue of Somatic Mutations in Cancer, and their occurrence in hematologic malignancy cases is variable (Table 2).
The prevalence of CHIP did not significantly increase with age, which is likely attributable to the uneven distribution of patients in age subgroups (Fig. 1A). DNMT3A mutations were the most commonly observed in both Aβ-positive and Aβ-negative groups (Fig. 1B), in line with findings in previous Korean reference studies [2, 18, 19]. Regarding the variant types, Aβ-positive patients tended to exhibit a higher prevalence of missense variants (62.5%) than of nonsense and splice site variants (33.3% each), although the difference was not statically significant (P=0.546, Fig. 1C). All CHIP variants detected were SNVs (Table 2), with the most frequent base changes being C:G>T:A in both Aβ-positive and Aβ-negative groups (Fig. 1D), which is consistent with findings by Savola, et al. [9], who studied CHIP in patients with rheumatoid arthritis. When DNA is replicated, the most frequent type of mutation that occurs involves C>T transversion, which is notably characteristic of mutations associated with aging [20]. Among the patients with CHIP, VAFs were not significantly different between the Aβ-positive and Aβ-negative groups (Fig. 1E; median VAF, 4.45% vs. 3.83%, P=0.361). Finally, to evaluate the risk of brain Aβ deposition, multivariate logistic regression analysis was performed using age, sex, presence of CHIP, and presence of the APOE ε4 allele as variables (Fig. 1F). Although CHIP was not associated with brain Aβ deposition (OR 0.85, 95% CI 0.13–7.29, P=0.686), the APOE ε4 allele emerged as an independent risk factor for brain Aβ deposition (OR 6.58, 95% CI 1.70–33.80, P=0.011). Collectively, our results indicate that the characteristic patterns of CHIP in older patients with cognitive impairment reflect the aging of the hematopoietic system, characterized by C>T transitions and mutations in DNMT3A, a representative epigenetic modifier.
Our study had some limitations. First, this was a preliminary study investigating the relationship between CHIP and brain Aβ deposition; therefore, a power calculation for the sample size was not performed. Consequently, the statistical power may have been insufficient to detect the influence of CHIP on brain Aβ deposition. Second, the small sample size did not allow analysis of the association of CHIP with brain Aβ deposition according to gene, mutation type, and clone size. Third, the association between CHIP and brain Aβ deposition was studied cross-sectionally, and temporal causality was not evaluated.
In summary, older Korean patients with cognitive impairment had CHIP with typical age-related patterns, irrespective of the presence or absence of brain Aβ deposition, but CHIP was not found to be correlated with brain Aβ deposition. To our knowledge, this study was the first to investigate the relationship between CHIP and brain Aβ deposition in Korean patients using amyloid PET. However, our findings are preliminary, and a larger patient cohort is required to draw conclusive evidence. The protective effect of CHIP against AD may result from the migration of marrow-derived mutant cells into brain microglia by enhancing microglial phagocytosis [10]. Further research is required to understand the functional implications of CHIP on brain Aβ deposition, particularly in older individuals, as both CHIP and brain Aβ deposition are age-related phenomena.
Notes
AUTHOR CONTRIBUTIONS
Kim HR contributed to the study conception and design, and reviewed and revised the manuscript. Yun J was responsible for investigating clinical data, analyzing the results, performing statistical analysis, and drafting the manuscript. Youn YC assisted in collecting clinical samples. All authors have read and approved the final manuscript.
Appendix
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.3343/alm.2024.0086
References
1. Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. 2015; Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 126:9–16. DOI: 10.1182/blood-2015-03-631747. PMID: 25931582. PMCID: PMC4624443.
2. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. 2014; Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 371:2488–98. DOI: 10.1056/NEJMoa1408617. PMID: 25426837. PMCID: PMC4306669.
3. Jaiswal S, Libby P. 2020; Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 17:137–44. DOI: 10.1038/s41569-019-0247-5. PMID: 31406340. PMCID: PMC9448847.
4. Kim M, Kim JJ, Lee ST, Shim Y, Lee H, Bae S, et al. 2024; Association between aortic valve sclerosis and clonal hematopoiesis of indeterminate potential. Ann Lab Med. 44:279–88. DOI: 10.3343/alm.2023.0268. PMID: 38205526. PMCID: PMC10813825.
5. Mas-Peiro S, Hoffmann J, Fichtlscherer S, Dorsheimer L, Rieger MA, Dimmeler S, et al. 2020; Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J. 41:933–9. DOI: 10.1093/eurheartj/ehz591. PMID: 31504400. PMCID: PMC7033916.
6. Dorsheimer L, Assmus B, Rasper T, Ortmann CA, Ecke A, Abou-El-Ardat K, et al. 2019; Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 4:25–33. DOI: 10.1001/jamacardio.2018.3965. PMID: 30566180. PMCID: PMC6439691.
7. Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, et al. 2017; DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 130:753–62. DOI: 10.1182/blood-2017-04-777029. PMID: 28655780.
8. Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. 2017; Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 130:742–52. DOI: 10.1182/blood-2017-02-769869. PMID: 28483762. PMCID: PMC5553576.
9. Savola P, Lundgren S, Keränen MAI, Almusa H, Ellonen P, Leirisalo-Repo M, et al. 2018; Clonal hematopoiesis in patients with rheumatoid arthritis. Blood Cancer J. 8:69. DOI: 10.1038/s41408-018-0107-2. PMID: 30061683. PMCID: PMC6066480.
10. Bouzid H, Belk JA, Jan M, Qi Y, Sarnowski C, Wirth S, et al. 2023; Clonal hematopoiesis is associated with protection from Alzheimer's disease. Nat Med. 29:1662–70. DOI: 10.1038/s41591-023-02397-2. PMID: 37322115. PMCID: PMC10353941.
11. U.S. Food & Drug Administration. Drugs@FDA: FDA-approved drugs. Amyvid/labels for NDA202008. https://www.accessdata.fda.gov/drugsatfda_docs/label/2012/202008s000lbl.pdf. Updated on April 2012.
12. U.S. Food & Drug Administration. Drugs@FDA: FDA-approved drugs. Neuraceq/labels for NDA204677. https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/204677s000lbl.pdf. Updated on March 2014.
13. U.S. Food & Drug Administration. Drugs@FDA: FDA-approved drugs. Vizamyl/labels for NDA203137. https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/203137s000lbl.pdf. Updated on October 2013.
14. Sabri O, Seibyl J, Rowe C, Barthel H. 2015; Beta-amyloid imaging with florbetaben. Clin Transl Imaging. 3:13–26. DOI: 10.1007/s40336-015-0102-6. PMID: 25741488. PMCID: PMC4339690.
15. Jessen F, Amariglio RE, van Boxtel M, Breteler M, Ceccaldi M, Chételat G, et al. 2014; A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer's disease. Alzheimers Dement. 10:844–52. DOI: 10.1016/j.jalz.2014.01.001. PMID: 24798886. PMCID: PMC4317324.
16. Petersen RC. 2011; Clinical practice. Mild cognitive impairment. N Engl J Med. 364:2227–34. DOI: 10.1056/NEJMcp0910237. PMID: 21651394.
17. American Psychiatric Association. 2022. Diagnostic and statistical manual of mental disorders. 5th ed., text revision. American Psychiatric Association;Washington, DC: DOI: 10.1176/appi.books.9780890425787.
18. Bolton KL, Koh Y, Foote MB, Im H, Jee J, Sun CH, et al. 2021; Clonal hematopoiesis is associated with risk of severe Covid-19. Nat Commun. 12:5975. DOI: 10.1038/s41467-021-26138-6. PMID: 34645798. PMCID: PMC8514469. PMID: 3cb397d07d014cc5b6487816e9888d83.
19. Moon I, Kong MG, Ji YS, Kim SH, Park SK, Suh J, et al. 2023; Clinical, mutational, and transcriptomic characteristics in elderly Korean individuals with clonal hematopoiesis driver mutations. Ann Lab Med. 43:145–52. DOI: 10.3343/alm.2023.43.2.145. PMID: 36281508. PMCID: PMC9618905.
20. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. 2013; Signatures of mutational processes in human cancer. Nature. 500:415–21. DOI: 10.1038/nature12477. PMID: 23945592. PMCID: PMC3776390.
Fig. 1
Characteristics of CHIP variants in the Aβ-positive (N=43) and Aβ-negative (N=15) groups and forest plot of multivariate regression for the risk of brain Aβ deposition. (A) CHIP prevalence according to age. Bars indicate CHIP prevalence in different age groups. The line graph represents the cumulative prevalence of CHIP across the age groups. The absolute number of patients with CHIP and the total number of patients in each age group are displayed at the base of the graph. (B) CHIP prevalence according to gene. (C) Proportions of variant types in CHIP variants. (D) Percentages of different SNVs in CHIP variants. (E) VAFs of CHIP variants detected in the Aβ-positive and Aβ-negative groups. (F) Forest plot of multivariate logistic regression for the risk of brain Aβ deposition. Covariates included age, sex, presence of CHIP, and presence of the APOE ε4 allele.
Abbreviations: Aβ, β-amyloid; CHIP, clonal hematopoiesis of indeterminate potential; CI, confidence interval; OR, odds ratio; SNV, single-nucleotide variant; VAF, variant allele frequency.
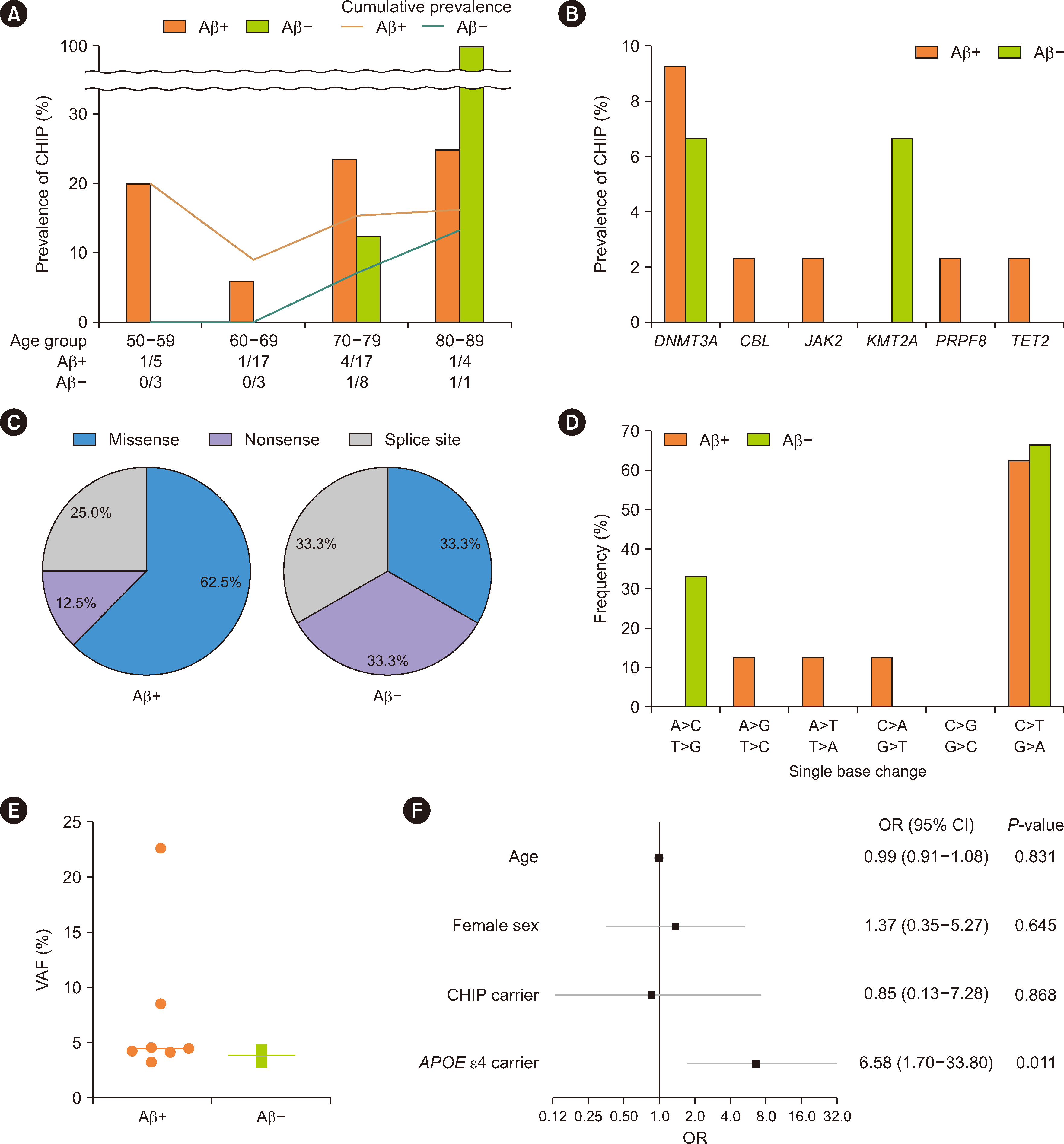
Table 1
Demographics of the study population
Table 2
CHIP variants observed in our study population
| Aβ group | Sex/age, yrs | Cognitive status | Gene and reference sequence | Coding DNA sequence and amino acid change | VAF (%)/total depth | COSMIC | |
|---|---|---|---|---|---|---|---|
| No. | Hematologic malignancy case count* | ||||||
| Aβ-positive | F/58 | Dementia | DNMT3A NM_022552 | c.1015-1G>A | 4.5/1,549 | None | None |
| F/69 | Dementia | DNMT3A NM_022552 | c.2206C>T (p.Arg736Cys) | 4.1/1,941 | COSM231560 | 18 | |
| M/72 | MCI | PRPF8 NM_006445 | c.6427C>T (p.Arg2143Cys) | 8.5/1,735 | None | None | |
| F/74 | Dementia | TET2 NM_001127208 | c.1588C>T (p.Gln530Ter) | 4.6/1,602 | COSM4383816 | 3 | |
| F/77 | SCD | DNMT3A NM_022552 | c.1040T>A (p.Leu347Gln) | 3.5/1,764 | None | None | |
| JAK2 NM_004972 | c.1849G>T (p.Val617Phe) | 22.6/1,683 | COSM12600 | 42,776 | |||
| F/79 | Dementia | DNMT3A NM_022552 | c.2478+1G>A | 4.2/851 | COSM4766079 | 1 | |
| F/80 | MCI | CBL NM_005188 | c.1253T>C (p.Phe418Ser) | 3.3/2,000 | COSM34070 | 8 | |
| Aβ-negative | M/73 | Dementia | KMT2A NM_001197104 | c.2329T>G (p.Ser777Ala) | 3.3/1,997 | None | None |
| F/80 | MCI | DNMT3A NM_022552 | c.1851+1G>A | 4.4/1,477 | COSM5878805 | 1 | |
| c.2311C>T (p.Arg771Ter) | 3.2/1,811 | COSM231563 | 15 | ||||




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download