Abstract
Background
Droplet digital (dd)PCR is a new-generation PCR technique with high precision and sensitivity; however, the positive and negative droplets are not always effectively separated because of the “rain” phenomenon. We aimed to develop a practical optimization and evaluation process for the ddPCR assay and to apply it to the detection of BRAF V600E in fine-needle aspiration (FNA) specimens of thyroid nodules, as an example.
Methods
We optimized seven ddPCR parameters that can affect “rain.” Analytical and clinical performance were analyzed based on histological diagnosis after thyroidectomy using a consecutive prospective series of 242 FNA specimens.
Results
The annealing time and temperature, number of PCR cycles, and primer and probe concentrations were found to be more important considerations for assay optimization than the denaturation time and ramp rate. The limit of blank and 95% limit of detection were 0% and 0.027%, respectively. The sensitivity of ddPCR for histological papillary thyroid carcinoma (PTC) was 82.4% (95% confidence interval [CI], 73.6%–89.2%). The pooled sensitivity of BRAF V600E in FNA specimens for histological PTC was 78.6% (95% CI, 75.9%–81.2%, I2=60.6%).
Conclusions
We present a practical approach for optimizing ddPCR parameters that affect the separation of positive and negative droplets to reduce rain. Our approach to optimizing ddPCR parameters can be expanded to general ddPCR assays for specific mutations in clinical laboratories. The highly sensitive ddPCR can compensate for uncertainty in cytological diagnosis by detecting low levels of BRAF V600E.
Droplet digital (dd)PCR is a new-generation PCR technique with high precision and sensitivity that allows absolute quantification of nucleic acid target sequences [1-3]. Compared with conventional PCR and quantitative (q)PCR, ddPCR uses emulsion chemistry to partition the specimens into approximately 20,000 nL-sized droplets that contain mutant-type DNA (positive) or wild-type DNA (negative) before amplification [4, 5]. After the PCR, the positive and negative droplets are divided based on a fluorescence threshold, and the positive droplets are quantified using Poisson statistics, which provide absolute quantification of nucleic acids without the need for a standard curve [5-7].
Although ddPCR has many advantages, the positive and negative droplets are not always effectively separated. Droplets exhibiting fluorescence falling between positive and negative are termed “rain” [6, 8]. The cause of rain is unclear, but it can interfere with threshold setting and lead to false-negative or false-positive results. To minimize rain, the factors that may affect ddPCR require optimization [8, 9]. We applied ddPCR to the detection of BRAF V600E in cell lines as an example to evaluate various options for establishing ddPCR assays with minimal rain in clinical laboratories. We considered seven ddPCR assay parameters, including the denaturation time, annealing time and temperature, number of PCR cycles, ramp rate, and primer and probe concentrations. Finally, we evaluated the analytical performance of the optimized BRAF V600E assay.
As ddPCR is highly sensitive, ddPCR results of clinical specimens are regarded as final confirmation. However, careful consideration of the clinical application of a molecular genetic assay is essential, particularly in identifying the specific purpose for which the assay is used. In this study, we analyzed the clinical performance of ddPCR targeting BRAF V600E in fine-needle aspiration (FNA) specimens. Thyroid tissue specimens from ultrasound-guided FNA are widely used to diagnose thyroid cancer [10]. FNA provides a cytological diagnosis of a malignant or benign nodule for preoperative determination. The cytological diagnosis of benign thyroid lesions is accurate. In contrast, the risk of malignancy of indeterminate thyroid nodules, such as atypia of undetermined significance/follicular lesion of undermined significance (AUS/FLUS) and follicular neoplasm/suspicious for follicular neoplasm (FN/SFN), is 11%–23% and 20%–29%, respectively [11]. Therefore, additional molecular assays are used to investigate these indeterminate nodules [10, 12-14].
Papillary thyroid carcinoma (PTC) accounts for approximately 80% of all thyroid cancers, and BRAF V600E is one of the most common oncogenic mutations detected in PTC [15]. This mutation activates continuous intracellular signaling (via the MAPK pathway), resulting in abnormal cell proliferation and increased tumor cell survival [16, 17]. Molecular methods for detecting the BRAF V600E mutation in FNA specimens need to be sensitive for accurate detection, and they should undergo thorough validation before adoption in clinical laboratories because of the generally low number of mutation-carrying cells present in FNA samples. Clinical performance analysis of molecular genetics assays in FNA specimens requires comparison with histological diagnosis post-thyroidectomy rather than cytological results. [18].
In this study, we assessed the clinical performance of the optimized BRAF V600E assay for papillary thyroid carcinoma (PTC) confirmed by histology after thyroidectomy using consecutive prospective FNA specimens. We previously studied various methods (e.g., sequencing, real-time PCR) to detect BRAF V600E in clinical FNA specimens and reported the diagnostic rates of these methods for thyroid cancer [19-21]. Therefore, we compared the clinical performance of three molecular methods (real-time (q)PCR, mutant enrichment with 3′-modified oligonucleotide [MEMO] sequencing, and ddPCR). Furthermore, we conducted a meta-analysis of the molecular diagnostic rate of BRAF V600E in FNA specimens for thyroid cancer in a single institution with the same clinical environment and system.
This study was approved by the Institutional Review Board of Samsung Medical Center (Seoul, Korea). To evaluate the clinical performance of the optimized BRAF V600E assay, clinical specimens meeting both of the following criteria were selected: 1) specimens obtained by FNA from patients suspected of having thyroid cancer based on ultrasound examination, 2) FNA samples that had undergone BRAF genetic testing using both qPCR and MEMO sequencing methods for clinical diagnosis, and 3) sufficient remaining DNA regardless of the genetic test results. Ultimately, a total of 242 specimens meeting these criteria were consecutively collected during the study period.
We optimized seven ddPCR parameters for the detection of BRAF V600E, including the denaturation time, annealing and temperature, number of PCR cycles, ramp rate, and primer and probe concentrations (Table 1). The initial ddPCR experimental conditions were set per the manufacturer’s instructions for the ddPCR Supermix for Probe (no dUTP) (https://www.bio-rad.com/sites/default/files/webroot/web/pdf/lsr/literature/10026868.pdf, last accessed on December 12, 2022), as follows: 1) the thermal cycles consisted of one cycle at 95°C for 10 mins, 40 cycles at 94°C for 30 secs and at 60°C for 1 min, one cycle at 98°C for 10 mins, and hold at 4°C; 2) BRAF V600E target primers and probes (Supplemental Data Table S1) were designed according to the Droplet Digital PCR Applications Guideline (https://dna.uga.edu/wp-content/uploads/sites/51/2021/03/ddPCR-Application-Guide.pdf, last accessed on December 13, 2022) and used at final concentrations of 500 nM and 250 nM, respectively; 3) the ramping rate was set to 2°C/sec. We used 30 ng of DNA extracted from K-562 cells (ATCC, Manassas, VA, USA) and SK-MEL-28 cells (ATCC), which harbor wild-type BRAF and a homozygous BRAF V600E mutation, respectively. Droplets were generated using the QX200 system (Bio-Rad, Hercules, CA, USA), and the data were analyzed using QuantaSoft (v1.7.4. 0917) (Bio-Rad).
The limit of blank (LOB) and limit of detection (LOD) were evaluated according to CLSI guideline EP17A2 [22]. The LOB was evaluated under the following conditions: 1 instrument×1 reagent lot×1 blank specimen (distilled water)×32 replicates per specimen×3 days×1 operator=96 total replicates, and was calculated using the rank position:
The LOD was evaluated under the following conditions: 1 instrument×1 reagent lot×7 low-concentration specimens×7 replicates per specimen×3 days×1 operator=147 total replicates. The seven low-concentration specimens comprised 1%, 0.5%, 0.1%, 0.05%, 0.01%, 0.005%, and 0.001% of the BRAF V600E mutation, and the LOD and 95% confidence interval (CI) were calculated using probit analysis.
ddPCR intra-assay performance was evaluated based on the CV (%) in five low-concentration specimens (1%, 0.5%, 0.1%, 0.05%, and 0.01% of the BRAF V600E mutation), and was calculated as follows:
The precision SD was analyzed following CLSI guideline EP05-A2 [23] and was calculated as follows:
where B is the SD of the daily means, N is the number of replicates per run, and Sr2 is the repeatability variance estimate. The data were visualized using R (v3.6.0).
Experienced radiologists performed FNA with ultrasound guidance. FNA specimens were smeared onto glass slides and fixed in 95% alcohol for Papanicolaou and hematoxylin and eosin staining for cytological examination. The remaining materials were used for BRAF V600E mutation analysis using molecular assays, including 1) ddPCR, 2) a clamping qPCR assay with a GENEDIA BRAF gene mutation detection kit (Green Cross Medical Science, Yongin, Korea), and 3) MEMO sequencing [19].
Cytological diagnoses for all FNA specimens were reviewed according to the Bethesda System for Reporting Thyroid Cytopathology [24]. In cases where patients underwent subsequent thyroidectomy because of suspected clinically malignant tumors, the histological diagnosis and clinical pathological features were reviewed.
To analyze clinical performance, cases of BRAF V600E in FNA and histologically diagnosed as PTC were considered true positive. Cases of BRAF V600E in FNA and not histologically diagnosed as PTC were considered false positives. Cases in which BRAF V600E in FNA was negative but histologically diagnosed as PTC were considered false negative. Cases in which BRAF V600E was negative, and thyroid tissue was pathologically benign were considered true negative. As it is difficult to perform surgery on benign nodules, cases in which BRAF V600E was not detected by all molecular methods (ddPCR, qPCR, and MEMO sequencing) and that were cytologically benign were classified as true negative. Kappa inter-rater agreement values with 95% CIs were calculated using MedCalc software v11.5.1.0 (MedCalc, Ostend, Belgium) to compare the BRAF V600E mutation detection rate of ddPCR with those of qPCR and MEMO sequencing. Statistical analyses were performed using SPSS Statistics version 20.0.0 (SPSS Inc., Chicago, IL, USA) and the statistical software package R, version 4.2.0.
The optimized conditions for detecting the BRAF V600E mutation were as follows: a denaturation time of 1 min 30 secs, annealing time of 1 min 30 secs, annealing temperature of 57°C, 40 PCR cycles, ramp rate of 2°C/sec, primer concentration of 750 nM, and probe concentration of 250 nM (Fig. 1 and Supplemental Data Fig. S1). A comparison of the data obtained with the standard and optimized protocols using FNA specimens is presented in Supplemental Data Fig. S2.
Under the optimized ddPCR conditions, the LOB was evaluated in 96 replicates. The BRAF V600E mutation was detected six times (0.05, 0.06, 0.06, 0.06, 0.16, and 0.32 copies/µL). The blank results were not normally distributed; therefore, the nonparametric method (rank order) was used to calculate the LOB at 95% confidence. The rank was calculated to be 91.7, and the LOB was 0% (value at rank position 92). The LOD was tested in 147 replicates of seven low-concentration specimens. According to probit analysis, the 95% LOD was 0.027% (95% CI, 0.016%–0.076%).
We evaluated precision by testing duplicate specimens per run and five different batches. The CV value, based on the percentage of mutation (log10), was calculated using linear regression analysis; R2=0.953 (y=–24.877x+4.948) (Supplemental Data Fig. S3 and Table S2). The results showed that a lower mutation burden was associated with a higher CV.
One hundred and two cases that were histologically diagnosed as PTC were determined to be true positives and 62 cases, including histologically benign cases (N=8) and cytologically benign cases negative for the three molecular methods (N=54), were determined to be true negatives (Fig. 2). The PTC detection rates (sensitivity) of ddPCR, MEMO sequencing, and qPCR in any thyroid nodules were 82.4% (95% CI, 73.6%–89.2%), 76.5% (95% CI, 67.0%–84.3%), and 65.7% (95% CI, 55.6%–74.8%), respectively (Fig. 3). Discordant results among the three molecular assays are described in Supplemental Data Table S3. The PTC detection rates of ddPCR, MEMO sequencing, and qPCR in indeterminate nodules were 75.7% (95% CI, 56.5%–89.7%), 65.5% (95% CI, 45.7%–82.1%), and 55.2% (95% CI, 35.7%–73.6%), respectively. The diagnostic sensitivity of cytological malignancy for PTC was 65.7% (95% CI, 55.6%–74.8%), whereas the sensitivity of combined results (malignancy or suspicious for malignant cells [SMC]) increased to 84.3% (95% CI, 75.8%–90.8%). The combined sensitivity of ddPCR and cytological malignancy was 86.3% (95% CI, 78.0%–92.3%). The combined sensitivity of ddPCR and cytological malignancy or SMC increased to 92.2% (95% CI, 85.1%–96.6%). There was no significant association between the BRAF V600E mutation status in FNA specimens and clinicopathological features of PTC (Supplemental Data Table S4).
A meta-analysis of the clinical sensitivity of BRAF V600E in FNA specimens for PTC diagnosis was performed considering previous studies [19-21] conducted in a single institution (Samsung Medical Center, Seoul, Korea). The pooled sensitivity of the BRAF V600E mutation in the four studies (including our study, 990 FNA specimens) based on a univariate random-effects meta-analysis was 78.6% (95% CI, 75.9%–81.2%, I2=60.6%).
To adopt ddPCR in clinical diagnostics, optimization of the ddPCR parameters and validation of its analytical performance are crucial to ensure sensitive and accurate results (Fig. 1). Maier, et al. [9] optimized the quenchers and Witte, et al. [8] optimized the number of chemical parameters, including the MgCl2, dNTP, and polymerase concentrations, to improve the separation of positive and negative droplets. According to the Digital MIQE Guidelines, the annealing temperature, number of PCR cycles, and primer and probe concentrations are essential factors for assay optimization [25, 26]. Our experimental findings corroborated that the annealing time and temperature, number of PCR cycles, and primer and probe concentrations are more important factors in minimizing rain than the other factors we considered (Fig. 1 and Supplemental Data Fig. S1).
We analyzed whether low-concentration mutations can be detected by ddPCR for BRAF V600E under the optimized conditions. The LOD was 0.027%, and the LOB was zero, indicating a high sensitivity. According to internal evaluation data, the 95% LOD of qPCR was 0.598%. This value was calculated by probit analysis using eight replicates of six low-concentration specimens (1.5%, 1.0%, 0.5%, 0.1%, 0.05%, and 0.01%). The detection sensitivity of MEMO sequencing of the BRAF V600E mutation was 0.1% in FNA specimens [19]. When measuring low-concentration mutations, determining the maintenance of reproducibility is crucial. ddPCR for BRAF V600E showed difficulty in maintaining a 30% CV at low concentrations <0.05% variant allele frequency.
The initial experiment using the standard protocol and clinical specimens failed to adequately separate positive and negative droplets for establishing the threshold line (Supplemental Data Fig. S2), hampering the comparison of the clinical performance between the standard and optimized protocols with clinical specimens to determine any significant differences in the detection rates. When clinical sensitivity was determined based on PTC cases diagnosed after thyroidectomy, ddPCR showed the highest clinical sensitivity among ddPCR, MEMO sequencing, and qPCR. Interestingly, among the three molecular assays, only ddPCR detected BRAF V600E, and six cases were later histologically determined to be PTCs (Fig. 2). Some cases were preoperatively cytologically diagnosed as benign (N=1), AUS/FLUS (N=2), FN/SFN (N=1), or malignant (N=2). Particularly among the cases that were not cytologically malignant (benign or indeterminate nodules) but histologically determined as PTCs, BRAF V600E was detected by the ddPCR molecular assay in 68.6% (Fig. 2): cytologically benign 33.3% (2/6), AUS/FLUS 62.5% (5/8), FN/SFN 50% (1/2), and SMC 84% (16/19). Therefore, the combination of cytological examination and molecular assays enhances clinical sensitivity. We recommend that in cases of indeterminate cytology results, a BRAF V600E mutation assay should be conducted using FNA specimens of thyroid nodules. Additional research is required, but the BRAF V600E mutation assay may aid in the diagnosis of PTC in cases where malignancy is suspected on ultrasonography but not confirmed by cytology [19, 20]. Kappa inter-rater agreement values with 95% CIs were calculated to compare the BRAF V600E mutation detection rates of ddPCR, qPCR, and MEMO sequencing. The κ values between ddPCR and qPCR and between ddPCR and MEMO sequencing were 0.788 (95% CI, 0.711–0.865) and 0.740 (95% CI, 0.665–0.824), respectively.
The pooled sensitivity of BRAF V600E for PTC in our meta-analysis (78.6%; 95% CI, 75.9%–81.2%) was higher than that (69.0%, 95% CI, 61%–76%) in a previous meta-analysis [27]. Although the number of FNA specimens in our study (990 FNA specimens from four studies) was lower than that in the previous study (9,924 FNA specimens from 47 studies), we reason that the bias may have been lower in our analysis because the studies included in our analysis were conducted in the same medical environment. Therefore, consistent result interpretation was possible.
According to previous studies [28-31], several clinicopathological features were significantly associated with the BRAF V600E mutation (e.g., tumor size, lymph node metastasis, extra-thyroidal extension, higher tumor, node, metastasis stage, and recurrence). However, our study had certain limitations. First, it did not reveal a significant association between clinicopathological features and the BRAF V600E mutation (Supplemental Data Table S4). As we analyzed the association between BRAF V600E in FNA specimens and clinical outcomes of PTC, there was a time difference between diagnosis and thyroidectomy. Furthermore, only 18 of the 102 cases with PTC were BRAF V600E-negative, making statistical comparison difficult. As the sensitivity of a molecular test increases, the occurrence of negative results decreases; therefore, the correlation between clinicopathological features and the BRAF V600E mutation may vary depending on the assay method used.
In conclusion, we presented a practical approach for optimizing ddPCR parameters (major and minor factors) that affect the separation of positive and negative droplets to reduce rain. This approach can be expanded to general ddPCR assays for specific mutations in clinical laboratories. ddPCR is a highly sensitive and valuable diagnostic tool to detect low levels of the BRAF V600E mutation and to play an important role in the decision for early thyroid cancer surgery. Therefore, ddPCR in FNA specimens can complement the uncertainty of cytological diagnoses, such as indeterminate nodules, and caution must be exercised even when nodules are diagnosed as benign.
Notes
AUTHOR CONTRIBUTIONS
Min YK and Park KS designed and carried out most of the experiments, and analyzed the data. Park KS and Kim JW conceived the study, designed the experiments, analyzed the data, and wrote the manuscript together with Min YK and Kim JK. All authors contributed to the manuscript and approved the submitted version.
Appendix
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.3343/alm.2023.0405
References
1. Huggett JF, Cowen S, Foy CA. 2015; Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 61:79–88. DOI: 10.1373/clinchem.2014.221366. PMID: 25338683.
2. Sreejith KR, Ooi CH, Jin J, Dao DV, Nguyen NT. 2018; Digital polymerase chain reaction technology - recent advances and future perspectives. Lab Chip. 18:3717–32. DOI: 10.1039/C8LC00990B. PMID: 30402632.
3. Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, et al. 2017; Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron. 90:459–74. DOI: 10.1016/j.bios.2016.09.082. PMID: 27818047.
4. Taylor SC, Carbonneau J, Shelton DN, Boivin G. 2015; Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for quantification of oseltamivir-resistant subpopulations. J Virol Methods. 224:58–66. DOI: 10.1016/j.jviromet.2015.08.014. PMID: 26315318.
5. Jones M, Williams J, Gärtner K, Phillips R, Hurst J, Frater J. 2014; Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, 'definetherain'. J Virol Methods. 202:46–53. DOI: 10.1016/j.jviromet.2014.02.020. PMID: 24598230. PMCID: PMC4003534.
6. Gerdes L, Iwobi A, Busch U, Pecoraro S. 2016; Optimization of digital droplet polymerase chain reaction for quantification of genetically modified organisms. Biomol Detect Quantif. 7:9–20. DOI: 10.1016/j.bdq.2015.12.003. PMID: 27077048. PMCID: PMC4827695.
7. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. 2011; High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 83:8604–10. DOI: 10.1021/ac202028g. PMID: 22035192. PMCID: PMC3216358.
8. Witte AK, Mester P, Fister S, Witte M, Schoder D, Rossmanith P. 2016; A systematic investigation of parameters influencing droplet rain in the Listeria monocytogenes prfA assay - reduction of ambiguous results in ddPCR. PLoS One. 11:e0168179. DOI: 10.1371/journal.pone.0168179. PMID: 27992475. PMCID: PMC5167268. PMID: 30f535116df4400e99868a27cff3b017.
9. Maier J, Lange T, Cross M, Wildenberger K, Niederwieser D, Franke GN. 2019; Optimized digital droplet PCR for BCR-ABL. J Mol Diagn. 21:27–37. DOI: 10.1016/j.jmoldx.2018.08.012. PMID: 30347270.
10. Xu X, Ma X, Zhang X, Cao G, Tang Y, Deng X, et al. 2019; Detection of BRAF V600E mutation in fine-needle aspiration fluid of papillary thyroid carcinoma by droplet digital PCR. Clin Chim Acta. 491:91–6. DOI: 10.1016/j.cca.2019.01.017. PMID: 30682328.
11. Lee ST, Kim SW, Ki CS, Jang JH, Shin JH, Oh YL, et al. 2012; Clinical implication of highly sensitive detection of the BRAF V600E mutation in fine-needle aspirations of thyroid nodules: a comparative analysis of three molecular assays in 4585 consecutive cases in a BRAF V600E mutation-prevalent area. J Clin Endocrinol Metab. 97:2299–306. DOI: 10.1210/jc.2011-3135. PMID: 22500044.
12. Park KS, Oh YL, Ki CS, Kim JW. 2015; Evaluation of the Real-Q BRAF V600E detection assay in fine-needle aspiration samples of thyroid nodules. J Mol Diagn. 17:431–7. DOI: 10.1016/j.jmoldx.2015.03.006. PMID: 25937618.
13. Choi R, Park KS, Kim JW, Ki CS. 2015; Evaluation of the Anyplex BRAF V600E real-time detection assay using dual-priming oligonucleotide technology in fine-needle aspirates of thyroid nodules. Ann Lab Med. 35:624–9. DOI: 10.3343/alm.2015.35.6.624. PMID: 26354351. PMCID: PMC4579107.
14. CLSI. 2020. Evaluation of detection capability for clinical laboratory measurement procedures; approved guideline. 2nd ed. Clinical and Laboratory Standards Institute;Wayne, PA: CLSI EP17-A2.
15. CLSI. 2004. Evaluation of precision performance of quantitative measurement methods; approved guideline. 2nd ed. Clinical and Laboratory Standards Institute;Wayne, PA: CLSI EP5-A2.
16. Cibas ES, Ali SZ. 2017; The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid. 27:1341–6. DOI: 10.1089/thy.2017.0500. PMID: 29091573.
17. Alexander EK, Cibas ES. 2022; Diagnosis of thyroid nodules. Lancet Diabetes Endocrinol. 10:533–9. DOI: 10.1016/S2213-8587(22)00101-2. PMID: 35752200.
18. Krauss EA, Mahon M, Fede JM, Zhang L. 2016; Application of the Bethesda classification for thyroid fine-needle aspiration: institutional experience and meta-analysis. Arch Pathol Lab Med. 140:1121–31. DOI: 10.5858/arpa.2015-0154-SA. PMID: 27684984.
19. Nikiforov YE, Ohori NP, Hodak SP, Carty SE, LeBeau SO, Ferris RL, et al. 2011; Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 96:3390–7. DOI: 10.1210/jc.2011-1469. PMID: 21880806. PMCID: PMC3205883.
20. Patel KN, Angell TE, Babiarz J, Barth NM, Blevins T, Duh QY, et al. 2018; Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg. 153:817–24. DOI: 10.1001/jamasurg.2018.1153. PMID: 29799911. PMCID: PMC6583881.
21. Steward DL, Carty SE, Sippel RS, Yang SP, Sosa JA, Sipos JA, et al. 2019; Performance of a multigene genomic classifier in thyroid nodules with indeterminate cytology: a prospective blinded multicenter study. JAMA Oncol. 5:204–12. DOI: 10.1001/jamaoncol.2018.4616. PMID: 30419129. PMCID: PMC6439562.
22. Cancer Genome Atlas Network. 2014; Integrated genomic characterization of papillary thyroid carcinoma. Cell. 159:676–90. DOI: 10.1016/j.cell.2014.09.050. PMID: 25417114. PMCID: PMC4243044.
23. Nikiforov YE, Nikiforova MN. 2011; Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 7:569–80. DOI: 10.1038/nrendo.2011.142. PMID: 21878896.
24. Xing M. 2013; Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–99. DOI: 10.1038/nrc3431. PMID: 23429735. PMCID: PMC3791171.
25. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. 2013; The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 59:892–902. DOI: 10.1373/clinchem.2013.206375. PMID: 23570709.
26. Huggett JF. dMIQE Group. 2020; The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin Chem. 66:1012–29. DOI: 10.1093/clinchem/hvaa125. PMID: 32746458.
27. Fnais N, Soobiah C, Al-Qahtani K, Hamid JS, Perrier L, Straus SE, et al. 2015; Diagnostic value of fine needle aspiration BRAF(V600E) mutation analysis in papillary thyroid cancer: a systematic review and meta-analysis. Hum Pathol. 46:1443–54. DOI: 10.1016/j.humpath.2015.06.001. PMID: 26232865.
28. Li C, Lee KC, Schneider EB, Zeiger MA. 2012; BRAF V600E mutation and its association with clinicopathological features of papillary thyroid cancer: a meta-analysis. J Clin Endocrinol Metab. 97:4559–70. DOI: 10.1210/jc.2012-2104. PMID: 23055546. PMCID: PMC3513529.
29. Liu C, Chen T, Liu Z. 2016; Associations between BRAF(V600E) and prognostic factors and poor outcomes in papillary thyroid carcinoma: a meta-analysis. World J Surg Oncol. 14:241. DOI: 10.1186/s12957-016-0979-1. PMID: 27600854. PMCID: PMC5012084.
30. Moon S, Song YS, Kim YA, Lim JA, Cho SW, Moon JH, et al. 2017; Effects of coexistent BRAFV600E and TERT promoter mutations on poor clinical outcomes in papillary thyroid cancer: a meta-analysis. Thyroid. 27:651–60. DOI: 10.1089/thy.2016.0350. PMID: 28181854.
31. Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, et al. 2015; Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol. 33:42–50. DOI: 10.1200/JCO.2014.56.8253. PMID: 25332244. PMCID: PMC4268252.
Fig. 1
Comparison of the standard and optimized ddPCR protocols. The standard and optimized ddPCR protocols were compared using mixtures of SK-MEL-28 cells (mutant BRAF V600E) and K-562 cells (wild-type BRAF V600). The variant allele frequencies in the mixtures were 5% and 10%. In the optimized ddPCR assay, negative (gray or dark dots) and positive droplets (blue dots represent BRAF V600E, green dots represent wild-type BRAF V600, and orange dots represent both positive dots [mutant and wild-type]) are clearly separated. (A and C) Initial protocol and 5% variant allele frequency. (B and D) Optimized protocol and 5% variant allele frequency. (E and G) Initial protocol and 10% variant allele frequency. (F and H) Optimized protocol and 10% variant allele frequency.
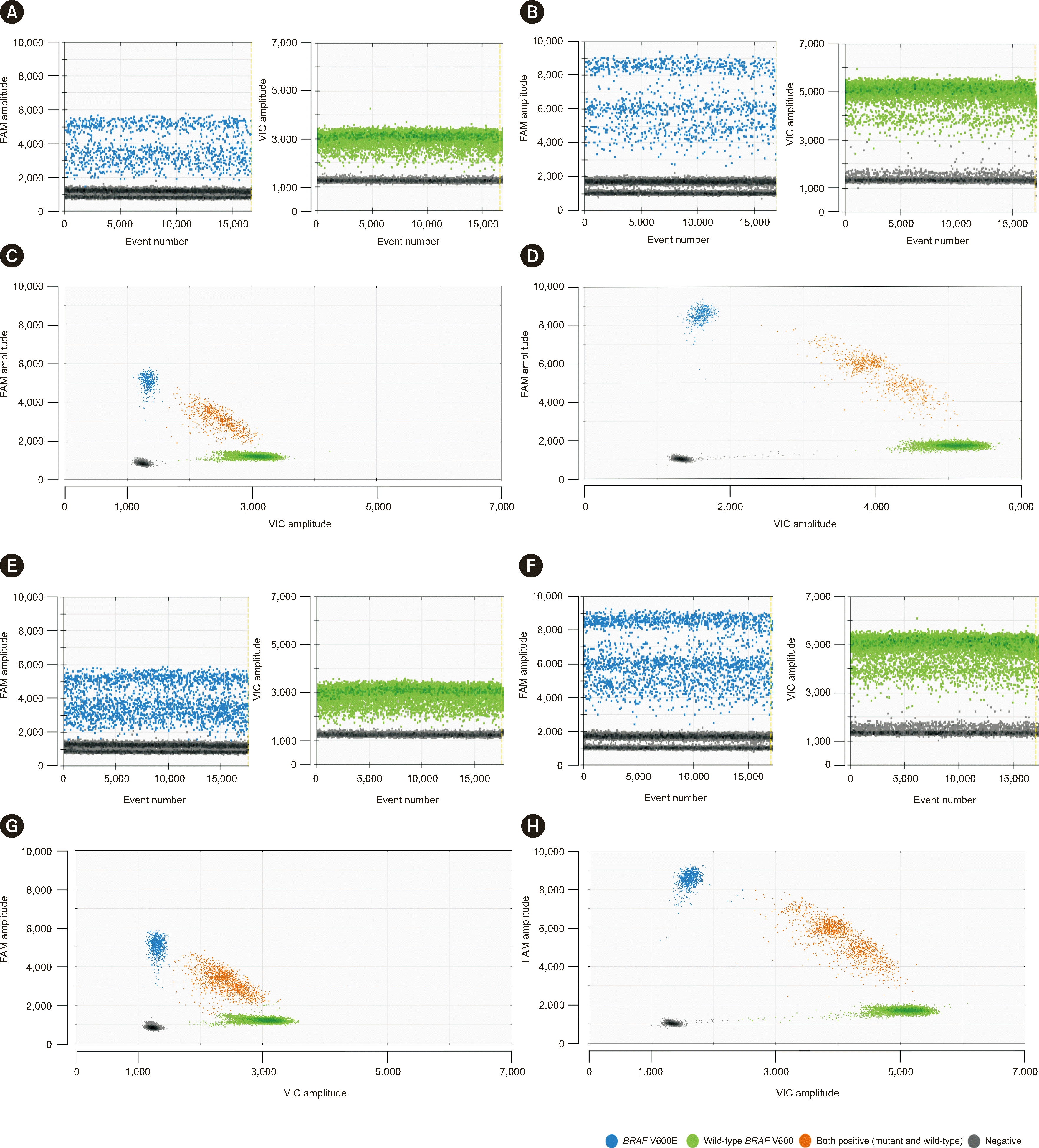
Fig. 2
Correlations among cytological diagnosis, results of the three molecular methods used for detecting the BRAF V600E mutation, and histological diagnosis. Benign nodules were operated in eight cases, including nodular hyperplasia (N=5), follicular adenoma (N=1), and Hürthle cell adenoma (N=2). One case was diagnosed as an indeterminate lesion.
Abbreviations: MEMO, mutant enrichment with 3′-modified oligonucleotide; ddPCR, droplet digital PCR; AUS/FLUS, atypia of undermined significance/follicular lesion of undetermined significance; FN/SFN, follicular neoplasm/suspicious for follicular neoplasm; SMC, suspicious for malignant cells; PTC, papillary thyroid carcinoma; MTC, medullary thyroid carcinoma.
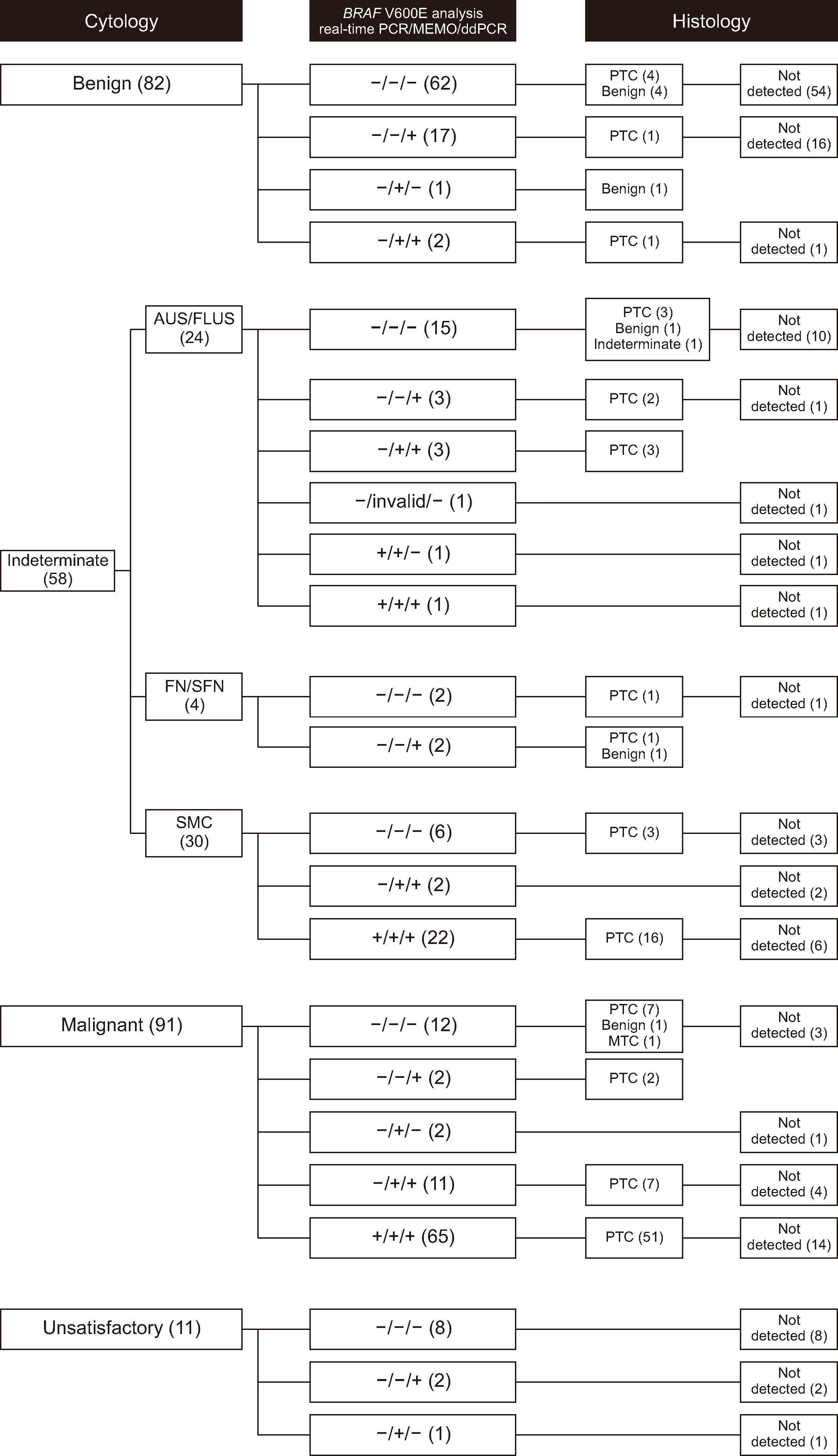
Fig. 3
Clinical performance of the molecular methods and cytology for PTC. The specificity and PPV of the cytological diagnosis of malignancy and the three molecular methods for detecting the BRAF V600E mutation in PTC were 100% (95% CI, 95.2%–100%) and 100%, respectively. Analytical performance was calculated as follows: sensitivity=true positive/(true positive+false negative)×100; specificity=true negative/(true negative+false positive)×100; PPV=true positive/(true positive+false positive)×100; NPV=true negative/(true negative+false negative)×100.
Abbreviations: PTC, papillary thyroid carcinoma; ddPCR, droplet digital PCR; NPV, negative predictive value; PPV, positive predictive value; SMC, suspicious for malignant cells.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download