Abstract
Tissue-specific adult stem cells are pivotal in maintaining tissue homeostasis, especially in the rapidly renewing intestinal epithelium. At the heart of this process are leucine-rich repeat-containing G protein-coupled receptor 5-expressing crypt base columnar cells (CBCs) that differentiate into various intestinal epithelial cells. However, while these CBCs are vital for tissue turnover, they are vulnerable to cytotoxic agents. Recent advances indicate that alternative stem cell sources drive the epithelial regeneration post-injury. Techniques like lineage tracing and single-cell RNA sequencing, combined with in vitro organoid systems, highlight the remarkable cellular adaptability of the intestinal epithelium during repair. These regenerative responses are mediated by the reactivation of conserved stem cells, predominantly quiescent stem cells and revival stem cells. With focus on these cells, this review unpacks underlying mechanisms governing intestinal regeneration and explores their potential clinical applications.
Tissue homeostasis is maintained throughout an organism’s lifespan by adult stem cells, whose self-renewal and differentiation processes are intricately regulated. The steady-state turnover rate of the intestinal epithelium is remarkably higher than other tissues and leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)+ crypt base columnar cells (CBCs) are identified as bona fide intes-tinal stem cells (ISCs) that replenish the gut epithelial lining (1, 2). In the mouse small intestine, crypt-bottom residing CBCs give rise to transit-amplifying (TA) cells that migrate toward the luminal side, where they stochastically differentiate into either absorptive- or secretory progenitors, leading to the generation of defined intestinal epithelial cells (IECs) including enterocytes, Paneth cells, goblet cells and enteroendocrine cells (3). Since these matured cells at the tips of the apical part keep shedding into the lumen via anoikis, a detachment-associated apoptotic death, the fine balance between self-renewal potential and the differentiation capacity of actively cycling CBCs is pivotal for intestinal integrity. In specific, the stem cell niche, composed of neighboring Paneth- and stromal cells as well as the surrounding microenvironment, provides CBCs with a range of molecules that modulate key signaling involved in stem cell fate, including the Wnt/R-spondin, Notch, bone morphogenetic protein (BMP), transforming growth factor (TGF)β, and epidermal growth factor (EGF) pathways (4, 5). Furthermore, along the crypt-TA zone-villus architecture axis, various cell-to-cell interactions between epithelial- and non-epithelial cells, extracellular matrix (ECM)-associated features (composition and stiffness), and even microbiota and their metabolites play crucial roles in regulating intestinal homeostasis (6-8). Building on this knowledge, Sato et al. (9) have reported that crypt bottom regions bearing ISCs and Paneth cells of mouse small intestine can be effectively cultured in vitro with the supplement of stromal niche factors and the ECM analog Matrigel, leading to the emergence of self-organized three-dimensional (3D) epithelial structures termed intestinal organoids. Currently, organoid culture technologies have been well-established for various conditions, such as origin (small intestine vs. colon), cell type (embryonic stem cells vs. adult ISCs), and species (humans and other animals), to support the expansion and differentiation of conventional LGR5+ ISCs in vitro.
Up to date, emerging evidence suggests that the regenerative responses upon injury in the intestinal epithe-lium are distinct from its homeostatic processes. Exposure to cytotoxic agents including chemicals and inflammatory cytokines disrupts crypt structure and abolishes LGR5+ ISCs in the intestine. CBCs are highly vulnerable to DNA damage mainly due to their active cell cycle with enhanced Wnt activity (10, 11) and Olfm4, one of the representative markers for CBCs, is rapidly down-regulated upon irradiation (10, 12). Moreover, a mouse model with diphtheria toxin-induced genetic ablation of LGR5+ stem cells showed no significant defect in intestinal homeostasis (13). Given the effective repair of the intestinal epithelium despite the absence of LGR5+ ISCs, and their subsequent resurgence implying the restoration of homeostasis, it is evident that alternative stem cell pools instead of CBCs drive the intestinal regenerative process. Lineage tracing of IECs in transgenic animal models and single-cell RNA sequencing-based identification of novel cell populations with an enhanced understanding of cell-cell communications, as well as mechanistic validation studies using the organoid system (14, 15), collectively highlight the pronounced cellular plasticity of the damaged intestinal epithelium during wound healing and regeneration. The core regenerative mechanisms can be broadly divided into two: (1) reactivation of LGR5− or LGR5low-conserved stem cells and (2) dedifferentiation of precursors and differentiated IECs into stem cell-like cells. In this review, we predominantly focus on the former category, particularly label-retaining, quiescent stem cells and revival stem cells (RSCs), highlighting their key characteristics and regulatory mechanisms with a brief summary of representative findings based on animal models and human studies. In addition, this review will introduce preliminary research on efforts to induce these regenerative-specific ISC populations for potential clinical applications.
CBCs continuously proliferate and undergo asymmetric division to differentiate, leading to the formation of crypt-villus ribbons in clonal labeling experiments (16, 17). In contrast, Potten et al. (18) reported a subset of cells in the crypt resided at the +4 position above the Paneth cells, demonstrating long-term retention of DNA labels. Compared to CBCs, these slow-cycling cells maintain their dormant state in homeostatic conditions, while they reactivate and exhibit both self-renewal and multipotency to regenerate intestinal epithelium in the absence of CBCs, implying that label-retaining cells (LRCs) represent a ‘reserved’ stem cell population (19). Based on further lineage tracing results, several candidates have been proposed as LRC markers including B cell-specific moloney murine leukemia virus integration site 1 (Bmi1), HOP homeobox (Hopx), SRY-box transcription factor 9 (SOX9), leucine-rich repeats and immunoglobulin-like domains 1 (Lrig1), mouse telomerase reverse transcriptase (mTert) and mex-3 RNA binding family member A (Table 1) (20-25); however, the molecular characteristics of reported LRCs might differ from each other and warrant detailed investigation. In terms of marker expression, for instance, Bmi1 and Hopx, initially identified as a marker for LRCs, are not exclusively expressed in +4 cells but also found in subpopulation of CBCs and even differentiated cells (26-28). In the intes-tinal epithelium of Sox9-eGFP mice, crypt cells could be categorized into four populations based on Sox9 expre-ssion level (29). Notably, Sox9high cells that demonstrate label-retaining capacity and radioresistance exhibited gene signatures resembling both +4 cells and secretory progenitors. Roth et al. (30) identified Mmp7 and Defa1-expressing long-lived Paneth cells as the LRC population using an in vivo pulse-chase experiment with H2B-GFP mice for nuclear labeling. Meanwhile, fluorescence-based reporter assay revealed that Lgr5high cells exhibited substantial levels of transcripts associated with proposed quiescent cell markers including Bmi1, Hopx, Lrig1, and mTert as comparable levels to Hopx- or Bmi1 expressing cells (26, 31). Collectively, LRCs are perceived as a slow-cycling heterogeneous population predominantly located around the +4 position of the crypt base. They display a range of markers that overlap with those of CBCs and precursor cells, particularly those of the secretory lineage.
Despite the ongoing disputes regarding the detailed characteristics, a major criterion for identifying quiescent stem cells is their capacity to reactivate and lead the regeneration process upon injury. Interestingly, they also partici-pate in maintaining homeostasis. Lineage-tracing experiments under homeostatic conditions have shown that Bmi1- or Hopx expressing cells can generate clonal strips covering the crypt-villus axis as CBCs (20, 24, 32). Sangiorgi and Capecchi (20) verified the impact of Bmi1+ cell loss by tamoxifen injection to Bmi1Cre-ERT; Rosa26DTA mice and found that ablation of Bmi1 expression in the intestine resulted in marked weight loss and acute death of mice. In the histological analysis, crypt structures throughout the intestinal tract were remarkably disoriented and damaged but gradually replaced by intact ones over approximately a year.
In addition, several works suggest that secretory precursor cells are reserved as LRCs in homeostatic conditions. A lone-term pulse-chasing study of crypt cells using cytochrome p450 1a1-H2B-YFP mice revealed a novel LRC population that expresses secretory precursor markers ChgA and ChgB instead of conventional +4 cell markers such as Lrig1 and Hopx (33). Li et al. (34) further demonstrated that LRCs marked by H2B-GFP differ from Hopx-CreERT2 expressing quiescence stem cells, although both exhibit a decreased Wnt signaling activity than CBCs. H2B-GFP labeling conducted at various periods revealed two LRC populations: short-term-LRCs with enteroendoc-rine cell-like features could generate organoids in vitro, while long-term-LRCs exhibited Paneth cell traits but lack organoid-forming capacity. Of note, both ‘secretory’ LRCs are distinctive to Hopx-positive cells, which demonstrate a profound regenerative potential against radiation damage. On the contrary, the emergence of GFP-labeled cells at the +4 position in Bmi1-GFP mice is dependent on Atoh1 expression (35), which is one of the crucial transcription factors in secretory lineage specification. Indeed, RNA sequencing and trajectory analysis indicated that Bmi1GFP cells correlated to enteroendocrine cell signature and expressed ‘multi-capable’ enteroendocrine cell markers including prospero homeobox 1 (Prox1) (32, 36). Similar to Bmi1-expressing cells, Prox1+ cells were capable of expansion and reconstruction of the crypt-villi structure of homeostatic- and irradiated intestinal epithelium. The LRC-like properties of the enteroendocrine lineage have been also confirmed in CreERT2 mice targeting neurogenic differentiation 1 (NeuroD1), a basic helix-loop-helix transcription factor essential for enteroendocrine cell differentiation, or tryptophan hydroxylase 1 (Tph1), a marker gene for enterochromaffin cells (37). In this work, NeuroD1+ cells co-expressing ChgA or Tph1 persisted at the +4 position for several weeks and occasionally generated differentiated cells or reverted back to CBCs in homeostatic conditions. The authors further showed that a majority of these cells were also labeled by Hopx, a representative marker for +4 cells, and HopX+/Tph1+ cells displayed superior organoid forming capacity compared to HopX−/Tph1+ or HopX+/Tph1− cells. Meanwhile, Higa et al. (38) indicated that cyclin-dependent kinase inhibitor p57 can label the quiescent ISC populations at the +4 position, which also expressed the classical marker Bmi1. The p57 deletion driven by the villin gene promoter reduced the 5-ethynyl-2’-deoxyuridine (EdU)-retention capability of Bmi1+ cells, implying its role in the maintenance of dormant stem cells. According to the single-cell sequencing results, p57high cells did not express proliferation-associated genes but were enriched with enteroendocrine cell markers, such as Rfx6 and ChgB, with LRC markers. These p57+ cells could function as an alternative ISC population for postinjury epithelial regeneration in 5-fluorouracil (5-FU)-treated mice. Shifting the focus to a correlation between LRCs and goblet cells, a recent study indicated that Krüppel-like factor 4 (Klf4), typically expressed in differentiated IECs, is also observed in some Bmi1-expressing crypt cells that co-express goblet cell marker Muc2 (39). The cell-type specific deletion of Klf4 resulted in increased cell proliferation of Bmi1+ cells but reduced the number of goblet cells in the villi. Intestinal organoids derived from single Bmi1-YFP+ cells generated Klf4/Muc2+ differentiated cells, while those from Klf4-deleted Bmi1-YFP+ cells demonstrated enhanced proliferation without Muc2+ cell differentiation. Therefore, differentiation potential towards goblet cell lineage as well as the proliferation capacity of Bmi1-expressing LRCs might be regulated by Klf4 during homeostasis.
While accumulated evidence suggests the role of LRCs in maintaining homeostasis, certain studies contend that, in the absence of damage, LRCs do not contribute the epithelial cell turnover as conventional ISCs (30, 33), partially due to their insufficient endogenous Wnt signaling activity (38). Moreover, when Lgr5-expressing CBCs were genetically ablated in conjunction with intense radiation injury (over 6 Gy), the recovery of the intestinal epithelium was significantly impeded (40). This challenges the notions of radioresistance of +4 cells and the dispensability of Lgr5+ cells in intestinal regeneration. Hence, to understand the complex properties of the LRC populations, further studies are required on their reactions to diverse stimuli and the underlying mechanisms that drive regeneration. In this regard, a recent work by Chaves-Pérez et al. (41) explored the protective mechanism of LRCs in overcoming radiation- mediated gastrointestinal syndrome (GIS). The authors observed that the endogenous levels of the unconventional prefoldin RPB5 interactor (URI), a chaperone essential for maintaining DNA integrity following damage (42), were altered in the crypt following abdominal irradiation (8 to 14 Gy); URI levels decreased post-irradiation but returned to normal range during the recovery phase, implying its role in regeneration. Indeed, villin+ cell-specific overexpression of URI levels in Knock-in mice could protect mice from GIS, while deletion of URI resulted in intestinal failure resembling GIS even under normal conditions. Interestingly, URI expression was found predominantly at the upper crypt, especially Lgr5low cells. Further genetic tracing of URI-expressing cells in URI–YFP mice indicated that YFP labeled radiotolerant, slow–cycling cells in the crypt and these URI+ LRCs exhibited a repopulating capacity in homeostatic status as well as irradiated epithelium. It was noted that reduced URI expression in LRCs prompted cell cycle reactivation and proliferation, leading to increased radiosensitivity and apoptosis. Mechanistically, URI in LRCs function as a gatekeeper for the Wnt/β-catenin signaling, a crucial pathway for self-renewal of CBCs, by suppressing the nuclear translocation of β-catenin via direct binding. These findings underscore the pivotal role of URI in preserving the defining attributes of LRCs, namely quiescence and radioresistance. Another study by Chen et al. (43) proposes an experimental approach to enhance the regenerative potential of Lgr5− LRCs against GIS. To control the cytotoxic impact of reactive oxygen species (ROS) during injury, the authors overexpressed one of the ROS scavengers, TP53-induced glycolysis and apoptosis regulator (TIGAR) (44), in a stem cell-specific manner. Interestingly, TIGAR upregulation in Bmi1-expressing cells, but not in Lgr5+ cells, significantly improved the survival of irradiated mice. Followed lineage tracing of TIGAR-overexpressed Bmi1+ cells showed that the proliferation of these cells was enhanced both in vivo and in vitro (confirmed by organoid culture). Transient overexpression of TIGAR via adenovirus in organoids also promoted the growth in Bmi1+ cell-derived organoids following irradiation, whereas organoids from Lgr5+ cells were not affected by TIGAR expression level. This beneficial impact of TIGAR was mediated by activating protein 1 (AP-1) activity, considering that administration of a specific inhibitor of AP-1, 3-PA, could reverse the enhanced recovery of Bmi1-specific TIGAR-overexpressed mice. These studies suggest novel insights into how various innate stress-regulating molecules can modulate the reactivation and regenerative response of LRCs.
The great advances in genetic engineering technologies have enabled the development of various mouse models to study cell type-specific behaviors, thereby facilitating in-depth investigations into the characteristics of both Lgr5+ ISCs and LRCs; however, research regarding human stem cells has been constrained due to experimental limitations. To overcome this challenge, Ishikawa et al. (45) conducted an elegant work exploring the dynamics of human colonic stem cells (hCoSCs) during homeostasis as well as injury status with hCoSC-implanted mouse models, generated as previously reported (46). Single-cell sequencing of human colonic epithelium revealed both LGR5+ stem cell and +4 cell-like secretory precursor populations. Notably, the LGR5+ stem cell subpopulation in humans lacks of proliferative markers, in contrast to that of mice. Visualization of LGR5 mRNA and Ki67 protein displayed Ki67−, slow-cycling LGR5+ cells were localized mainly at the crypt bottom. Based on the observation that p27, a negative regulator of the cell cycle, readily labels these non-dividing LGR5+ CoSCs, genetically engineered human colonoids bearing LGR5- and p27 reporters simultaneously were generated. In in vitro organoid culture, most of the LGR5+/p27+ cells were in the G0 status, while LGR5+/p27− cells expanded rapidly to differentiate. Within the orthotopically transplanted human colonoids, LGR5+/p27+ cells were localized at the de-novo crypt bottom, mirroring their position in human colonic tissue, and exhibited LRC-like behavior in the in vivo EdU label retention assay. Moreover, these cells were tolerable to chemotherapeutic 5-FU treatment; Indeed, 5-FU treatment ablated murine LGR5+ CBCs but enriched p27-expressing cells in human LGR5+ cells and progeny of LGR5+/p27+ cells generated stirps in 5-FU-treated injured lesion. Another intriguing point is the damage-induced dedifferenti-ation procedure of mature IECs in the presence of LGR5+/p27+ hCoSCs, suggesting that LGR5+ cells drive the regeneration process in the human intestine. This study supports previous findings that slow-cycling, quiescent stem cells contribute to the intestinal regeneration process. However, by revisiting the role of LGR5+ ISCs in this context, it also underscores the need for further research into pioneering specific markers for quiescent stem cells and understanding the mechanisms that govern their state of quiescence and cell cycle dynamics, especially in the light of interspecies variations.
Beyond the canonical niche cues, such as Wnt and BMP signaling cascades, the Hippo-yes-associated protein (YAP) signaling axis plays a pivotal role in the guidance of intestinal physiology from development to homeostasis (47). Accumulating evidence also suggests that the activation of the YAP signaling pathway orchestrates cell fate remodeling, crucial for complete tissue regeneration. YAP activation, characterized by nuclear translocation of YAP, is apparent in damaged intestinal epithelium from both mouse models and human tissues (12, 48-50), and suppression of YAP activity either with genetic ablation or inhibitory chemicals hinders the recovery of injured epithelium (12, 49). Regarding this phenomenon, Gregorieff et al. (12) demonstrated that rapid but transient activation of YAP signaling occurs in IECs in response to whole-body irradiation. YAP activation, in conjunction with EGF receptor signaling, led to a decline in canonical Wnt signaling, subsequently suppressing the expression of CBC markers Lgr5 and Olfm4 and Paneth cell differentiation in the crypt region.
To achieve an in-depth understanding of the regeneration-oriented cellular adaptation process regulated by YAP signaling, single-cell RNA sequencing on the repairing mouse intestine was performed to classify the radiation injury-affected IEC populations and identify damage-responsive subsets (51). Transcriptional profiling data of single cells showed profound differences in the IEC popula-tions between normal- and irradiated epithelium, including a loss of Lgr5+ CBCs and regional shift in mature cells to less-differentiated states as previously described. In the regenerative tissue, the authors identified the distinct and novel cell populations, characterized by the exclusive expression of DNA damage response and cell survival markers, including Clusterin (Clu) and Ly6a/Sca-1. As this subset of cells did not exhibit the proposed previous ISC markers associated with +4 cells as well as CBCs, they were designated as “revival stem cells” or RSCs, with Clu as their defining marker. Using BAC-Clu-GFP transgenic mice which marked endogenous Clu-expressing cells, authors revealed that Clu-GFP+ cells constituted a rare (less than 0.1%) population in the crypt, distinct from Lgr5 or Olfm4-expre-ssing CBCs. Using the TAM-induced ClutdTomato mice-based cell labeling technique, authors proposed that Clu+ cells were able to contribute to the homeostatic epithelial turnover, as evidenced by crypt-villus ribbon formation. Impo-rtantly, ribbon emergence correlated with the contribution of CLU+ progeny to Lgr5+ CBCs, as well as to differentiated lineages. In addition, the YAP1 gene signature was markedly enriched in the RSC population, and the emergence of Clu+ cells in the intestinal epithelium is regulated by a YAP1 activity-dependent manner. Moreover, RSCs exhibited the capability to reconstruct the injured crypt and compensate for the loss of residual CBCs, thereby replenishing the Crypt-villi structure during epithelial damage; Indeed, in ‘Cluless’ mice, intestine-specific Clu-deleted models by Tamoxifen treatment in CluDTA mice, exposures to irradiation or administration with the colitogen, dextran sodium sulfate (DSS), significantly reduced the length of intestine and crypt numbers due to the impaired regenerative capacity compared to controls. This is the first study to propose that RSCs, driven by YAP signaling, represent a slow-cycling, novel subset of ISCs essential for intestinal regeneration.
Of note, both prolonged activation and inactivation of YAP impair the growth of intestinal organoids; single-cell focused imaging and genomic analysis further indicated that YAP1 activity must exhibit spatial, temporary fluctuations between cells to drive Notch-DLL1 interaction-dependent symmetry breaking during normal organoid generation (52). These findings may also shed light on how the dynamic transition between regenerative (symmetric)- and homeostatic (asymmetric) states is regulated in the intestinal epithelium. The intervention of niche cellular components in this context is investigated in 2020 by Roulis et al. (53). They focused on the characterization of mesenchymal niche cells supporting epithelial homeostasis and turnover. In specific, rare pericryptal fibroblasts (RPPFs) expressing cyclooxygenase-2 (Cox-2) encoded by Ptgs2, named as RPPFs, mediated the adenoma formation in APCmin mice, implying their potential role in boosting the stemness of ISCs. Co-culture of organoids with RPPFs or prostaglandin E2 (PGE2), one of the Cox-2-generated prostaglandins, led to a transformation of spheroid-like structures. Interestingly, the budding-free, cyst-like morphology was a major characteristic of immature intestine-derived organoids from the embryonic period (54). These organoids consisted of undifferentiated, Wnt signaling-independent Lgr5low-cells and displayed a primitive fetal gene signature, including Sca-1 and yap target genes such as Clu, Ctgf, Ankrd1, and Anxa10 (48, 54). It is also reported that epithelial transcriptomes have to be reprogrammed into fetal colonic epithelial profiles via YAP activation to initiate the regenerative process (48). Similarly, RPPFs-derived PGE2 resulted in Sca-1+ RSC mobilization in the damaged epithelium by inducing YAP nuclear translocation. Furthermore, as observed in a YAP-deficient context, deletion of a PGE2 receptor Ptger4 in the epithelium significantly perturbed the normal repair response against irradiation, validating the PGE2–Ptger4–Yap signaling axis between intestinal epithelial-mesenchymal niche in RSC induction and regeneration. In addition, TGFβ1 also exert pleiotropic roles in this regenerative circumstances (55). Following irradiation-induced damage, the level of TGFβ1 noticeably surged around the injury site due to the action of damage-responsive macrophage. Surprisingly, addition of TGFβ1 to organoid culture led to fetal-like conversion characterized by RSC gene signature. Utilizing transwell-based intestinal organoid-stromal cell co-culture system, it is revealed that TGFβ1 could activate stromal cells, enhancing their ability to induce RSC population in organoids. Mechanistically, TGFβ1 led to an increase in chromatin accessibility in transcription factor motifs for Sox9, which is involved in YAP pathway. These findings provide a detailed depiction of how non-epithelial cells respond to intestinal epithelial injuries and their roles in contributing to regeneration, in terms of RSC induction.
Ectopic induction of the RSC population can be driven by an immune reaction against infection. As a murine patho-symbiont helminth in the intestine, Heligmosomoides polygyrus disrupts the epithelial barrier upon chronic infection (56), leading to a transformation of infected crypts into granuloma-associated phenotype (GAC) (57). Notably, GACs no longer expressed Lgr5 and Olfm4, suggesting the depletion of CBCs. Instead, infection with H. polygyrus upregulated the interferon (IFN) signaling-associated gene signature in GACs with the highest transcription of the Ly6a gene. An increase in IFN-γ, produced by stimulated T lymphocytes, augmented the Ly6a+ cells within the crypt while suppressing Lgr5 expression, even in the absence of infection. In addition, GACs underwent fetal-like reprogramming and GAC-derived organoids displayed a hyper-proliferative spheroid formation (54), suggesting that H. polygyrus infection and subsequent epithelial injury enhances the regenerative property of crypt region, marked by the emergence of RSCs. A similar study by Karo-Atar et al. (58) further elucidated that conditioned media from H. polygyrus bakeri (Hpb-CM) can mediate the Clu+ RSC induction in organoids, partially via the Hippo-YAP axis-dependent manner; however, the authors argued the role of IFN-γ on supporting the RSC-inducing effect of Hpb-CM, as the addition of IFN-γ to Hpb-CM-treated organoids reduced the Clu+ RSC populations despite the increment of Ly6a transcription. It is also found that the acquisition of the RSC phenotype following H. polygyrus infection was simultaneously accompanied by a reduced differentiation of secretory lineages, goblet cells and tuft cells, which play a crucial role in defending against helminth invasion. Moreover, type 2 immune response effectors, interleukin (IL)-4 and IL-13, could counteract the impact of Hpb-CM in RSC regulation. Overall, this study presents a new perspective on the interactions between infectious injury factors and the subsequent immune reaction in regulating RSC induction and epithelial reprogramming.
Although the intestinal epithelium is one of the most potent regenerative tissues in the body, transient and/or permanent defects in its repairing capacity often lead to detrimental outcomes, as observed in chemotherapy-induced diarrhea (CID) and radiotherapy-mediated GIS (59, 60). Given that Lgr5+ CBCs are rapidly depleted following injury, therapeutic strategies targeting alternative ISCs are imperative to enhance the endogenous regenerative poten-tial. In this context, reactivation of quiescent stem cells with cell cycle regulators could be a promising therapeutic approach. Palbociclib, a U.S. Food and Drug Administration-approved CDK4/6 inhibitor for treating breast cancer, has been reported to govern ISC fate, offering protection against chemotherapy-induced damage and promoting the recovery of the intestinal epithelium in CID models (60). By blocking cell cycle progression, Palbociclib reduced cell proliferation of Lgr5-expressing cells and protected them from 5-FU toxicity both in vitro and in vivo. Surprisingly, co-treatment of 5-FU and Palbociclib unexpectedly activated the expansion of Bmi1+ quiescent cells at the +4 position, implying an independent mechanism of action for Palbo-ciclib based on the cell cycle status.
Enhancement of the RSC induction offers beneficial effects on intestinal regeneration. A recently reported novel combination comprises eight components (8C) designed to mimic the authentic regenerative state of intestinal organoids (61). Compared to the original culture media, 8C led to a generation of hyper-proliferative organoids enriched for injury-associated regenerative phenotype. Within the composition, the epigenetic modulators, valproic acid (VPA) and EPZ6438, play critical roles in reprogramming IECs into RSC-like cells expressing Sca-1, Anxa1, Reg3b, and Clu. It is noted that a combined administration of VPA (HDAC inhibitor) and EPZ6438 (EZH2 inhibitor) facilitated intestinal epithelial healing with enhanced crypt reconstitution upon irradiation. Mechanistically, these two chemicals led to a global alteration in epigenetic modification patterns in injured IECs, especially inducing a down-regulation of H3K27me3 at promoter regions of YAP target genes. Furthermore, a potent YAP pathway inhibitor, verteporfin, significantly reverted the phenotype of VPA/EPZ6438-treated organoids, bringing them back to a normal state without regenerative signatures. Thus, YAP signaling might underlie the RSC-inducing capacity of VPA/EPZ6438. Meanwhile, to unravel the governing mechanisms that orchestrate intestinal homeostasis and regeneration, the Liberali group classified single-cell-derived organoids by phenotype into seven clusters following the treatment with a chemical library (62). Using the image-based organoid screening platform combined with hierarchical interaction score, the authors revealed that among various signaling pathways, the retinoid X receptor (RXR) pathway controls the transition between homeostatic- and regenerative status of IECs in conjunction with the YAP signaling. Treatment of RXR agonist (all-trans retinoic acid, atRA) or inhibitor (RXRi) to organoids altered cytoplasmic/nuclear YAP1 localization and subsequent symmetric breaking. In specific, RXRi-treated organoids acquired regenerative transcriptome patterns associated with YAP target genes and a fetal-like regenerative state, while organoids exposed to atRA underwent rapid maturation, which primarily consisted of enterocytes. Finally, in vivo administration of RXRi significantly alleviated the pathologic symptoms of the radiation-mediated colitis model, suggesting RXR antagonists as potent therapeutics for intestinal regeneration.
Transplantation of intestinal organoids can be beneficial to reconstitute the damaged intestinal epithelium. Given the positive role of TGFβ1 in RSC induction, Chen et al. (55) compared the therapeutic impact of TGFβ1-stimulated organoids to repair DSS-mediated injury with naïve organoids. After transplantation via rectal route, TGFβ1-treated organoids led to an improvement in histological- and pathological features of colitis in comparison to control organoids. The engraftment capacity of TGFβ1-treated organoids was also superior than counterparts in the co-transplantation assay, emphasizing the usefulness of TGFβ1 pretreatment in organoid-based cell therapy.
In addition to organoids, adult stem cells can be applied to treat intestinal disorders. In specific, mesenchymal stem cells (MSCs) serve as a primary source for stem cell therapy owing to their immunomodulatory functions and tissue-regenerative capacity. Based on previous works that under-scored the therapeutic benefits of naïve MSCs in the colitis model (63-65), several approaches have been explored to enhance their therapeutic efficacy. In 2021, we utilized MSC spheroids (MSC3D) for this purpose, given that 3D-cultured cells exhibit a potent secretory capacity compared to two-dimensional (2D)-grown counterparts (MSC2D) (66). After confirming the superior therapeutic impacts of MSC3D compared to MSC2D in alleviating the DSS-induced colitis model, we further investigated the phenotypic changes of IECs induced by MSCs with organoid assay. Interestingly, organoids cultured with either MSCs or their conditioned media (CM) underwent dramatic morphological transitions, shifting from a budding shape to a cystic spheroid, one of featured phenotypes indicative of fetal-like organoids enriched with RSCs. Indeed, these transformed organoids contained more abundant Ly6a+ cells than control organoids, and this observation was especially evident in organoids exposed to MSC3D or MSC3D-CM. Given the suspected involvement of paracrine factors in this phenomenon, we focused on PGE2, one of the major therapeutic mediators from MSCs (64) as well as a potent inducer of the RSC population in the intestinal epithelium (53), and measured its concentration in MSC-CM. It is found that MSC3D could produce a much higher level of PGE2 in comparison to MSC2D. In addition, blockage of the PGE2 action with chemical inhibitors successfully diminished the RSC-inducing impact of MSCs on organoids. Therefore, this study elucidates the role of MSCs as a source of the RSC-inducing factor, PGE2, illustrating how MSCs directly contribute to intestinal epithelial regeneration.
In summary, the intestinal epithelium contains a diverse and dynamic set of ‘potential’ or ‘reserve’ stem cells for compensation of homeostatic ISCs. In specific, the recently introduced concept of RSCs, along with their regulatory and inductive mechanisms, enriches our understanding of intestinal regeneration (Fig. 1); however, from both experimental and technical perspectives, numerous pitfalls exist in understanding the complexities of intestinal plasticity. Genetic differences in reporter alleles used in lineage labeling often lead to misinterpretations the cell fate tracing results for injury-specific ISC candidates (34). The myriad methodological approaches used to induce epithelial injury (i.e., irradiation, chemotherapeutics, colitogen and infectious agents), each with their distinct etiology, also pose challenges to the integrated understanding of the subsequent regenerative responses (67). Finally, the prono-unced cellular plasticity in the IECs during the continuous regenerative cascade complicates the classification and characterization of individual stem cell populations that drive intestinal repair. For instance, the characteristics of reported injury-responsive ISCs often manifest overlapping features, especially in terms of marker expression (Table 1). In most cases, +4 cells tend to be positive for secretory lineage markers (36). When quiescent p57+/Bmi1+ subsets reactivate upon injury, they undergo spatiotemporal reprogramming and start to express RSC-associated fetal markers (38). On the contrary, it has been reported that representative RSC markers, Clu and Ly6a, are differentially regulated among the RSC population (57, 58). Collectively, advanced lineage tracing strategies with specificity and refined genetic ablation techniques are needed for a comprehensive assessment of candidate markers, aiding the identification of reliable core markers for each regenerative ISCs.
Although YAP-dependent fetal-like reprogramming is crucial for cryptic RSC-driven regeneration, it has been also identified as playing an essential role in addressing villi injury scenarios. A poly(I:C)-mediated damage model in mice that mimics acute viral gastroenteritis demonstrates villi-specific loss with severe atrophy without any disturbance in the crypt region (68). During the regeneration of the villi structure, a specialized cell type referred to as atrophy-induced villus epithelial cells (aVECs) emerged to cover the atrophic villi lesion before the restoration of barrier junctions. Interestingly, aVECs lost their mature enterocyte markers but gained fetal markers Clu and Msln and expressed nuclear YAP protein, indicating that adaptive epithelial differentiation into reparative cells resembles a major feature of fetal conversion process as suggested previously (69). Another crucial consideration is the pote-ntial correlation between the reactivation of quiescent ISCs/RSCs and tumorigenesis. Indeed, uncontrolled activation of YAP pathway is regarded as an oncogenic driver (47), and several studies have reported the possible contribution of quiescent ISCs/RSCs in tumor initiation (12, 20, 53). Therefore, the fine balance required in therapeutic induction of quiescent ISCs/RSCs to avoid adverse effects and further research is needed on the upstream regulators that ensure YAP-mediated regenerative responses transition back to a homeostatic state instead of advancing toward tumorigenesis. Moreover, it offers insights for establishment of innovative therapeutic strategies to enhance the intrinsic repairing potential of the intestine.
Notes
References
1. Barker N, van Es JH, Kuipers J, et al. 2007; Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 449:1003–1007. DOI: 10.1038/nature06196. PMID: 17934449.
2. Janebodin K, Buranaphatthana W, Ieronimakis N, Hays AL, Reyes M. 2013; An in vitro culture system for long-term expansion of epithelial and mesenchymal salivary gland cells: role of TGF-β1 in salivary gland epithelial and mesenchymal differentiation. Biomed Res Int. 2013:815895. DOI: 10.1155/2013/815895. PMID: 23841093. PMCID: PMC3690740.
3. Gehart H, Clevers H. 2019; Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 16:19–34. DOI: 10.1038/s41575-018-0081-y. PMID: 30429586.
4. Sato T, van Es JH, Snippert HJ, et al. 2011; Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469:415–418. DOI: 10.1038/nature09637. PMID: 21113151. PMCID: PMC3547360.
5. de Lau W, Barker N, Low TY, et al. 2011; Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 476:293–297. DOI: 10.3410/f.12001958.14669072. PMID: 21727895.
6. Meran L, Baulies A, Li VSW. 2017; Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cells Int. 2017:7970385. DOI: 10.1155/2017/7970385. PMID: 28835755. PMCID: PMC5556610.
7. Shaker A, Rubin DC. 2010; Intestinal stem cells and epithelial-mesenchymal interactions in the crypt and stem cell niche. Transl Res. 156:180–187. DOI: 10.1016/j.trsl.2010.06.003. PMID: 20801415. PMCID: PMC3019104.
8. Wang D, Odle J, Liu Y. 2021; Metabolic regulation of intestinal stem cell homeostasis. Trends Cell Biol. 31:325–327. DOI: 10.1016/j.tcb.2021.02.001. PMID: 33648839.
9. Sato T, Vries RG, Snippert HJ, et al. 2009; Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. DOI: 10.3410/f.1158598.621586. PMID: 19329995.
10. Yan KS, Chia LA, Li X, et al. 2012; The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 109:466–471. DOI: 10.1073/pnas.1118857109. PMID: 22190486. PMCID: PMC3258636.
11. Tao S, Tang D, Morita Y, et al. 2017; Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 36:2920–2921. Erratum for: EMBO J 2015;34:624-640. DOI: 10.15252/embj.201490700. PMID: 25609789. PMCID: PMC4365032.
12. Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, Wrana JL. 2015; Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 526:715–718.
13. Tian H, Biehs B, Warming S, et al. 2011; A reserve stem cell population in small intestine renders Lgr5-positive cells dis-pensable. Nature. 478:255–259. DOI: 10.1038/nature10408. PMID: 21927002. PMCID: PMC4251967.
14. Ahn JS, Kang MJ, Seo Y, Kim HS. 2023; Intestinal organoids as advanced modeling platforms to study the role of host-microbiome interaction in homeostasis and disease. BMB Rep. 56:15–23. DOI: 10.5483/bmbrep.2022-0182. PMID: 36379514. PMCID: PMC9887104.
15. Choi WH, Bae DH, Yoo J. 2023; Current status and prospects of organoid-based regenerative medicine. BMB Rep. 56:10–14. DOI: 10.5483/bmbrep.2022-0195. PMID: 36523211. PMCID: PMC9887105.
16. Snippert HJ, van der Flier LG, Sato T, et al. 2010; Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 143:134–144. DOI: 10.3410/f.5964956.13138055. PMID: 20887898.
17. Winton DJ, Blount MA, Ponder BA. 1988; A clonal marker induced by mutation in mouse intestinal epithelium. Nature. 333:463–466. DOI: 10.1038/333463a0. PMID: 3163778.
18. Potten CS, Gandara R, Mahida YR, Loeffler M, Wright NA. 2009; The stem cells of small intestinal crypts: where are they? Cell Prolif. 42:731–750. DOI: 10.1111/j.1365-2184.2009.00642.x. PMID: 19788585. PMCID: PMC6496740.
19. Richmond CA, Shah MS, Carlone DL, Breault DT. 2016; An enduring role for quiescent stem cells. Dev Dyn. 245:718–726. DOI: 10.1002/dvdy.24416. PMID: 27153394. PMCID: PMC4912863.
20. Sangiorgi E, Capecchi MR. 2008; Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 40:915–920. DOI: 10.1038/ng.165. PMID: 18536716. PMCID: PMC2906135.
21. Barriga FM, Montagni E, Mana M, et al. 2017; Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell. 20:801–816.e7.
22. Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, Magness ST. 2015; SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroente-rology. 149:1553–1563.e10. DOI: 10.1053/j.gastro.2015.07.004. PMID: 26170137. PMCID: PMC4709179.
23. Powell AE, Wang Y, Li Y, et al. 2012; The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 149:146–158. DOI: 10.3410/f.717952209.793478796. PMID: 22464327. PMCID: PMC3563328.
24. Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. 2011; Interconversion between intestinal stem cell popula-tions in distinct niches. Science. 334:1420–1424. DOI: 10.1126/science.1213214. PMID: 22075725. PMCID: PMC3705713.
25. Montgomery RK, Carlone DL, Richmond CA, et al. 2011; Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 108:179–184. DOI: 10.3410/f.7984956.8361054. PMID: 21173232. PMCID: PMC3017192.
26. Muñoz J, Stange DE, Schepers AG, et al. 2012; The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31:3079–3091.
27. Itzkovitz S, Lyubimova A, Blat IC, et al. 2011; Single-molecule transcript counting of stem-cell markers in the mouse inte-stine. Nat Cell Biol. 14:106–114. DOI: 10.1038/ncb2384. PMID: 22119784. PMCID: PMC3292866.
28. López-Arribillaga E, Rodilla V, Pellegrinet L, et al. 2015; Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development. 142:41–50. DOI: 10.1242/dev.107714. PMID: 25480918.
29. Van Landeghem L, Santoro MA, Krebs AE, et al. 2012; Activa-tion of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 302:G1111–G1132. DOI: 10.1152/ajpgi.00519.2011. PMID: 22361729. PMCID: PMC3362093.
30. Roth S, Franken P, Sacchetti A, et al. 2012; Paneth cells in intestinal homeostasis and tissue injury. PLoS One. 7:e38965. DOI: 10.1371/journal.pone.0038965. PMID: 22745693. PMCID: PMC3380033.
31. Li N, Yousefi M, Nakauka-Ddamba A, et al. 2014; Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 3:876–891. DOI: 10.1016/j.stemcr.2014.09.011. PMID: 25418730. PMCID: PMC4235148.
32. Yan KS, Gevaert O, Zheng GXY, et al. 2017; Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 21:78–90.e6. DOI: 10.1016/j.stem.2017.06.014. PMID: 28686870. PMCID: PMC5642297.
33. Buczacki SJ, Zecchini HI, et al. Nicholson AM Intestinal label-retaining cells are secretory precursors expressing Lgr5. 2013; Nature. 495:65–69. DOI: 10.1016/j.stem.2017.06.014. PMID: 28686870. PMCID: PMC5642297.
34. Li N, Nakauka-Ddamba A, Tobias J, Jensen ST, Lengner CJ. 2016; Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastro-enterology. 151:298–310.e7. DOI: 10.1053/j.gastro.2016.04.049. PMID: 27237597. PMCID: PMC4961601.
35. Jadhav U, Saxena M, O’Neill NK, et al. 2017; Dynamic reorga-nization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell. 21:65–77.e5.
36. Engelstoft MS, Egerod KL, Lund ML, Schwartz TW. 2013; Enteroendocrine cell types revisited. Curr Opin Pharmacol. 13:912–921. DOI: 10.1016/j.coph.2013.09.018. PMID: 24140256.
37. Sei Y, Feng J, Samsel L, et al. 2018; Mature enteroendocrine cells contribute to basal and pathological stem cell dynamics in the small intestine. Am J Physiol Gastrointest Liver Physiol. 315:G495–G510. DOI: 10.1152/ajpgi.00036.2018. PMID: 29848020. PMCID: PMC6230697.
38. Higa T, Okita Y, Matsumoto A, et al. 2022; Spatiotemporal reprogramming of differentiated cells underlies regeneration and neoplasia in the intestinal epithelium. Nat Commun. 13:1500. DOI: 10.1038/s41467-022-29165-z. PMID: 35314700. PMCID: PMC8938507.
39. Katano T, Bialkowska AB, Yang VW. 2020; KLF4 regulates goblet cell differentiation in BMI1+ reserve intestinal stem cell lineage during homeostasis. Int J Stem Cells. 13:424–431. DOI: 10.15283/ijsc20048. PMID: 32840226. PMCID: PMC7691855.
40. Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. 2014; Lgr5+ stem cells are indispensable for radiation-induced intes-tinal regeneration. Cell Stem Cell. 14:149–159.
41. Chaves-Pérez A, Yilmaz M, Perna C, de la Rosa S, Djouder N. 2019; URI is required to maintain intestinal architecture during ionizing radiation. Science. 364:eaaq1165. DOI: 10.1126/science.aaq1165. PMID: 31147493.
42. Mita P, Savas JN, Briggs EM, et al. 2016; URI regulates KAP1 phosphorylation and transcriptional repression via PP2A phosphatase in prostate cancer cells. J Biol Chem. 291:25516–25528. DOI: 10.1074/jbc.m116.741660. PMID: 27780869. PMCID: PMC5207251.
43. Chen F, Zhang Y, Hu S, et al. 2020; TIGAR/AP-1 axis accelerates the division of Lgr5− reserve intestinal stem cells to reestablish intestinal architecture after lethal radiation. Cell Death Dis. 11:501. DOI: 10.1038/s41419-020-2715-6. PMCID: PMC7338449.
44. Lee P, Hock AK, Vousden KH, Cheung EC. 2015; p53- and p73-independent activation of TIGAR expression in vivo. Cell Death Dis. 6:e1842. DOI: 10.1038/cddis.2015.205. PMID: 26247727. PMCID: PMC4558498.
45. Ishikawa K, Sugimoto S, Oda M, et al. 2022; Identification of quiescent LGR5+ stem cells in the human colon. Gastroente-rology. 163:1391–1406.e24.
46. Sugimoto S, Ohta Y, Fujii M, et al. 2018; Reconstruction of the human colon epithelium in vivo. Cell Stem Cell. 22:171–176.e5. DOI: 10.1016/j.stem.2017.11.012. PMID: 29290616.
47. Seo Y, Park SY, Kim HS, Nam JS. 2020; The Hippo-YAP signaling as guardian in the pool of intestinal stem cells. Biome-dicines. 8:560. DOI: 10.3390/biomedicines8120560. PMID: 33271948. PMCID: PMC7760694.
48. Yui S, Azzolin L, Maimets M, et al. 2018; YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 22:35–49.e7. DOI: 10.1016/j.stem.2017.11.001. PMID: 29249464. PMCID: PMC5766831.
49. Kim HB, Kim M, Park YS, et al. 2017; Prostaglandin E2 activates YAP and a positive-signaling loop to promote colon rege-neration after colitis but also carcinogenesis in mice. Gas-troenterology. 152:616–630. DOI: 10.1053/j.gastro.2016.11.005. PMID: 27864128. PMCID: PMC5285392.
50. Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. 2010; The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24:2383–2388. DOI: 10.1101/gad.1978810. PMID: 21041407. PMCID: PMC2964748.
51. Ayyaz A, Kumar S, Sangiorgi B, et al. 2019; Single-cell transcri-ptomes of the regenerating intestine reveal a revival stem cell. Nature. 569:121–125. DOI: 10.1038/s41586-019-1154-y. PMID: 31019301.
52. Serra D, Mayr U, Boni A, et al. 2019; Self-organization and symmetry breaking in intestinal organoid development. Nature. 569:66–72. DOI: 10.1038/s41586-019-1146-y. PMID: 31019299. PMCID: PMC6544541.
53. Roulis M, Kaklamanos A, Schernthanner M, et al. 2020; Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 580:524–529. DOI: 10.1038/s41586-020-2166-3. PMID: 32322056. PMCID: PMC7490650.
54. Mustata RC, Vasile G, Fernandez-Vallone V, et al. 2013; Identifi-cation of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 5:421–432. DOI: 10.1016/j.celrep.2013.09.005. PMID: 24139799.
55. Chen L, Qiu X, Dupre A, et al. 2023; TGFB1 induces fetal reprogramming and enhances intestinal regeneration. Cell Stem Cell. 30:1520–1537.e8. DOI: 10.1016/j.stem.2023.09.015. PMID: 37865088. PMCID: PMC10841757.
56. Rapin A, Chuat A, Lebon L, Zaiss MM, Marsland BJ, Harris NL. 2020; Infection with a small intestinal helminth, Heligmosomoides polygyrus bakeri, consistently alters microbial communities throughout the murine small and large intestine. Int J Parasitol. 50:35–46. DOI: 10.1016/j.ijpara.2019.09.005. PMID: 31759944.
57. Nusse YM, Savage AK, Marangoni P, et al. 2018; Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 559:109–113. Erratum in: Nature 2018;562:E22. DOI: 10.1038/s41586-018-0257-1. PMID: 29950724. PMCID: PMC6042247.
58. Karo-Atar D, Ouladan S, Javkar T, et al. 2022; Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity. J Exp Med. 219:e20212311. DOI: 10.1084/jem.20212311. PMID: 35938990. PMCID: PMC9365672.
59. Stein A, Voigt W, Jordan K. 2010; Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based mana-gement. Ther Adv Med Oncol. 2:51–63. DOI: 10.1177/1758834009355164. PMID: 21789126. PMCID: PMC3126005.
60. Xiang J, Wang H, Tao Q, et al. 2023; CDK4/6 inhibitor modulating active and quiescent intestinal stem cells for prevention of chemotherapy-induced diarrhea. J Pathol. 260:235–247. DOI: 10.1002/path.6078. PMID: 36978197.
61. Qu M, Xiong L, Lyu Y, et al. 2021; Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res. 31:259–271. DOI: 10.1038/s41422-020-00453-x. PMID: 33420425. PMCID: PMC8027647.
62. Lukonin I, Serra D, Challet Meylan L, et al. 2020; Phenotypic landscape of intestinal organoid regeneration. Nature. 586:275–280. DOI: 10.1038/s41586-020-2776-9. PMID: 33029001. PMCID: PMC7116869.
63. Sala E, Genua M, Petti L, et al. 2015; Mesenchymal stem cells reduce colitis in mice via release of TSG6, independently of their localization to the intestine. Gastroenterology. 149:163–176.e20. DOI: 10.1053/j.gastro.2015.03.013. PMID: 25790743.
64. Kim HS, Shin TH, Lee BC, et al. 2013; Human umbilical cord blood mesenchymal stem cells reduce colitis in mice by activating NOD2 signaling to COX2. Gastroenterology. 145:1392–1403.e1-8. DOI: 10.1053/j.gastro.2013.08.033. PMID: 23973922.
65. González MA, Gonzalez-Rey E, Rico L, Büscher D, Delgado M. 2009; Adipose-derived mesenchymal stem cells alleviate expe-rimental colitis by inhibiting inflammatory and autoimmune responses. Gastroenterology. 136:978–989. DOI: 10.1053/j.gastro.2008.11.041. PMID: 19135996.
66. Regmi S, Seo Y, Ahn JS, et al. 2021; Heterospheroid formation improves therapeutic efficacy of mesenchymal stem cells in murine colitis through immunomodulation and epithelial regeneration. Biomaterials. 271:120752. DOI: 10.1016/j.biomaterials.2021.120752. PMID: 33730631.
67. Katsandegwaza B, Horsnell W, Smith K. 2022; Inflammatory bowel disease: a review of pre-clinical murine models of human disease. Int J Mol Sci. 23:9344. DOI: 10.3390/ijms23169344. PMID: 36012618. PMCID: PMC9409205.
68. Ohara TE, Colonna M, Stappenbeck TS. 2022; Adaptive differentiation promotes intestinal villus recovery. Dev Cell. 57:166–179.e6. DOI: 10.1016/j.devcel.2021.12.012. PMID: 35016013. PMCID: PMC9092613.
69. Miyoshi H, VanDussen KL, Malvin NP, et al. 2017; Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 36:5–24. DOI: 10.15252/embj.201694660. PMID: 27797821. PMCID: PMC5210160.
Fig. 1
Various modulators involved in revival stem cell (RSC) induction. Several endogenous (niche-associated)- and exogenous (helminth infection, chemical stimuli) signals can activate yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) pathway, a crucial inducer for fetal reprogramming and RSC appearance in intestinal epithelium. Hpb: Heligmosomoides polygyrus bakeri, Th: helper T cell, Tc: cytotoxic T cell, IFN: interferon, TGF: transforming growth factor, PGE2: prostaglandin E2, RPPFs: rare pericryptal fibroblasts, IL: interleukin, VPA: valproic acid, EPZ: EPZ6438, RXR: retinoid X receptor, Clu: Clusterin.
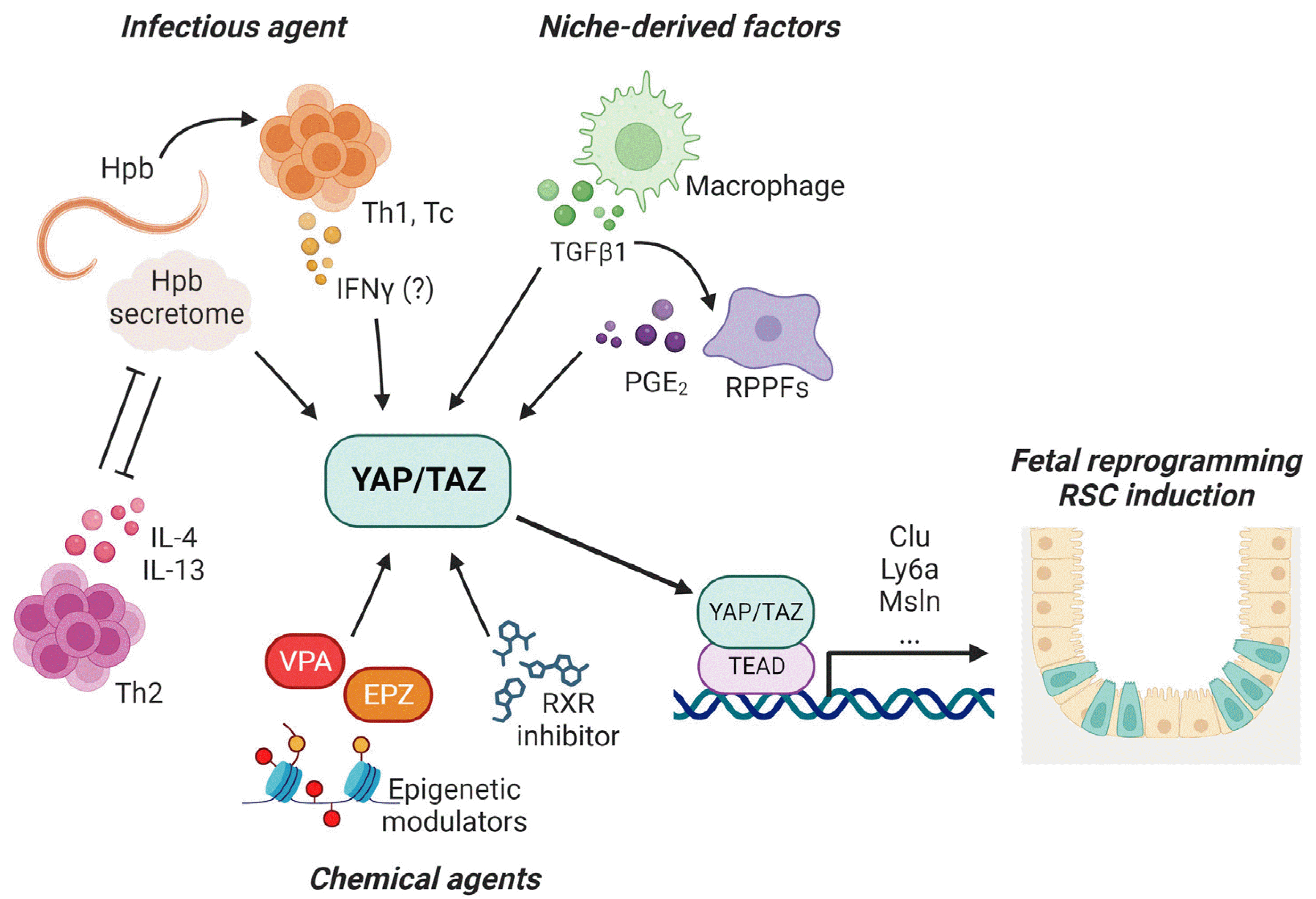
Table 1
Overview of key studies on quiescent stem cells
| Suggested markers | Used lineage tracing model | Position | Contribution | Reference | ||
|---|---|---|---|---|---|---|
|
|
||||||
| Homeostasis | Regeneration (damage type) | Tumorigenesis | ||||
| Bmi1 | Bmi1LacZ Bmi1DTA | +4, +5 | Y | Y (irradiation) | Y (inducer) | (20) |
| Mex3a | Mex3atdTomato | +1∼+4 | Y | Y (irradiation, 5-FU) | ND | (21) |
| Sox9 | Sox9GFP | +4 | N | Y (irradiation) | ND | (22) |
| Lrig1 | Lrig1LacZ | +1∼+5 | Y | Y (irradiation) | Y (repressor) | (23) |
| Hopx | HopxLacZ HopxmT-mG | +4 | Y | ND | ND | (24) |
| mTert | mTertGFP mTertLacZ | +4 | Y | Y (irradiation) | ND | (25) |
|
Bmi1 |
Bmi1LacZ Bmi1Confetti | +4/+5 | Y | ND | ND | (26) |
| Sox9 | Sox9GFP | +4/+5 | ND | Y (irradiation) | ND | (29) |
|
Bmi1 Prox1 |
Bmi1YFP Prox1GFP | +4 | N | Y (irradiation) | ND | (32) |
|
Bmi1 Hopx |
H2BGFP HopxtdTomato | +3∼+7 | N | Y (irradiation) | ND | (34) |
|
NeuroD1 Tph1 Hopx |
NeuroD1tdTomato Tph1tdTomato HopxtdTomato | +4 | Y | Y (irradiation) | ND | (37) |
|
p57 Bmi1 |
P57tdTomato Bmi1GFP | +4 | ND | Y (irradiation, Lgr5 ablation, 5-FU) | Y (inducer) | (38) |
Bmi1: B cell-specific Moloney murine leukemia virus integration site 1, Mex3a: mex-3 RNA binding family member A, Sox9: SRY-box transcription factor 9, Lrig1: leucine-rich repeats and immunoglobulin-like domains 1, Hopx: HOP homeobox, mTert: mouse telomerase reverse transcriptase, Prox1: prospero homeobox 1, NeuroD1: neurogenic differentiation 1, Tph1: tryptophan hydroxylase 1, Y: yes, N: no, ND: not determined, 5-FU: 5-fluorouracil, Lgr5: leucine-rich repeat-containing G protein-coupled receptor 5.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download