1. Baumann M, Krause M, Overgaard J, Debus J, Bentzen SM, Daartz J, et al. 2016; Radiation oncology in the era of precision medicine. Nat Rev Cancer. 16:234–249. DOI:
10.1038/nrc.2016.18. PMID:
27009394.

3. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. 2012; Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 366:883–892. Erratum in: N Engl J Med. 2012;367:976. DOI:
10.1056/NEJMoa1113205. PMID:
22397650. PMCID:
PMC4878653.

6. Brancato V, Cerrone M, Lavitrano M, Salvatore M, Cavaliere C. 2022; A systematic review of the current status and quality of radiomics for glioma differential diagnosis. Cancers (Basel). 14:2731. DOI:
10.3390/cancers14112731. PMID:
35681711. PMCID:
PMC9179305.

7. Zheng X, Yao Z, Huang Y, Yu Y, Wang Y, Liu Y, et al. 2020; Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun. 11:1236. Erratum in: Nat Commun. 2021;12:4370. DOI:
10.1038/s41467-020-15027-z. PMID:
32144248. PMCID:
PMC7060275.

8. Li G, Li L, Li Y, Qian Z, Wu F, He Y, et al. 2022; An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain. 145:1151–1161. DOI:
10.1093/brain/awab340. PMID:
35136934. PMCID:
PMC9050568.

9. Yu Y, Tan Y, Xie C, Hu Q, Ouyang J, Chen Y, et al. 2020; Development and validation of a preoperative magnetic resonance imaging radiomics-based signature to predict axillary lymph node metastasis and disease-free survival in patients with early-stage breast cancer. JAMA Netw Open. 3:e2028086. DOI:
10.1001/jamanetworkopen.2020.28086. PMID:
33289845. PMCID:
PMC7724560.

10. Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 2012; 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 30:1323–1341. DOI:
10.1016/j.mri.2012.05.001. PMID:
22770690. PMCID:
PMC3466397.

12. American Joint Committee on Cancer (AJCC). 2010. AJCC cancer staging manual. 7th ed. Springer;New York: p. 253–270. DOI:
10.1038/nrc.2016.18.
13. Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, et al. 2017; Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 44:1050–1062. DOI:
10.1002/mp.12123. PMID:
28112418. PMCID:
PMC5462462.
14. Larue RTHM, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, et al. 2017; Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 56:1544–1553. DOI:
10.1080/0284186X.2017.1351624. PMID:
28885084.

15. Velazquez ER, Parmar C, Jermoumi M, Mak RH, van Baardwijk A, Fennessy FM, et al. 2013; Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci Rep. 3:3529. DOI:
10.1038/srep03529. PMID:
24346241. PMCID:
PMC3866632.

16. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. 2017; Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77:e104–e107. DOI:
10.1158/0008-5472.CAN-17-0339. PMID:
29092951. PMCID:
PMC5672828.

17. Hastie T, Tibshirani R, Wainwright M. 2015. Statistical learning with sparsity: the Lasso and generalizations. CRC Press;New York: p. 2–10. DOI:
10.1201/b18401.
19. Li R, Xing L, Napel S, Rubin DL. 2019. Radiomics and radiogenomics: technical basis and clinical applications. CRC Press;Boca Raton: DOI:
10.1201/9781351208277.
20. Haralick RM, Shanmugam K, Dinstein IH. 1973; Textural features for image classification. IEEE Trans Syst Man Cybern. SMC-3:610–621. DOI:
10.1109/TSMC.1973.4309314.

22. Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. 2020; The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 295:328–338. DOI:
10.1148/radiol.2020191145. PMID:
32154773. PMCID:
PMC7193906.

23. Yan F, Kittler J, Mikolajczyk K, Tahir A. 2012; Non-sparse multiple kernel Fisher discriminant analysis. J Mach Learn Res. 13:607–642.
24. Park SH, Goo JM, Jo CH. 2004; Receiver operating characteristic (ROC) curve: practical review for radiologists. Korean J Radiol. 5:11–18. DOI:
10.3348/kjr.2004.5.1.11. PMID:
15064554. PMCID:
PMC2698108.

25. DeLong ER, DeLong DM, Clarke-Pearson DL. 1988; Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 44:837–845. DOI:
10.2307/2531595. PMID:
3203132.

26. MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. 2017; Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 284:228–243. DOI:
10.1148/radiol.2017161659. PMID:
28240562.

27. Honda O, Tsubamoto M, Inoue A, Johkoh T, Tomiyama N, Hamada S, et al. 2007; Pulmonary cavitary nodules on computed tomography: differentiation of malignancy and benignancy. J Comput Assist Tomogr. 31:943–949. DOI:
10.1097/RCT.0b013e3180415e20. PMID:
18043361.
28. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, et al. 2011; International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 6:244–285. DOI:
10.1097/JTO.0b013e318206a221. PMID:
21252716. PMCID:
PMC4513953.
29. Lim HJ, Ahn S, Lee KS, Han J, Shim YM, Woo S, et al. 2013; Persistent pure ground-glass opacity lung nodules ≥ 10 mm in diameter at CT scan: histopathologic comparisons and prognostic implications. Chest. 144:1291–1299. DOI:
10.1378/chest.12-2987. PMID:
23722583.

30. Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. 2017; Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 90:20160665. DOI:
10.1259/bjr.20160665. PMID:
27936886. PMCID:
PMC5685111.

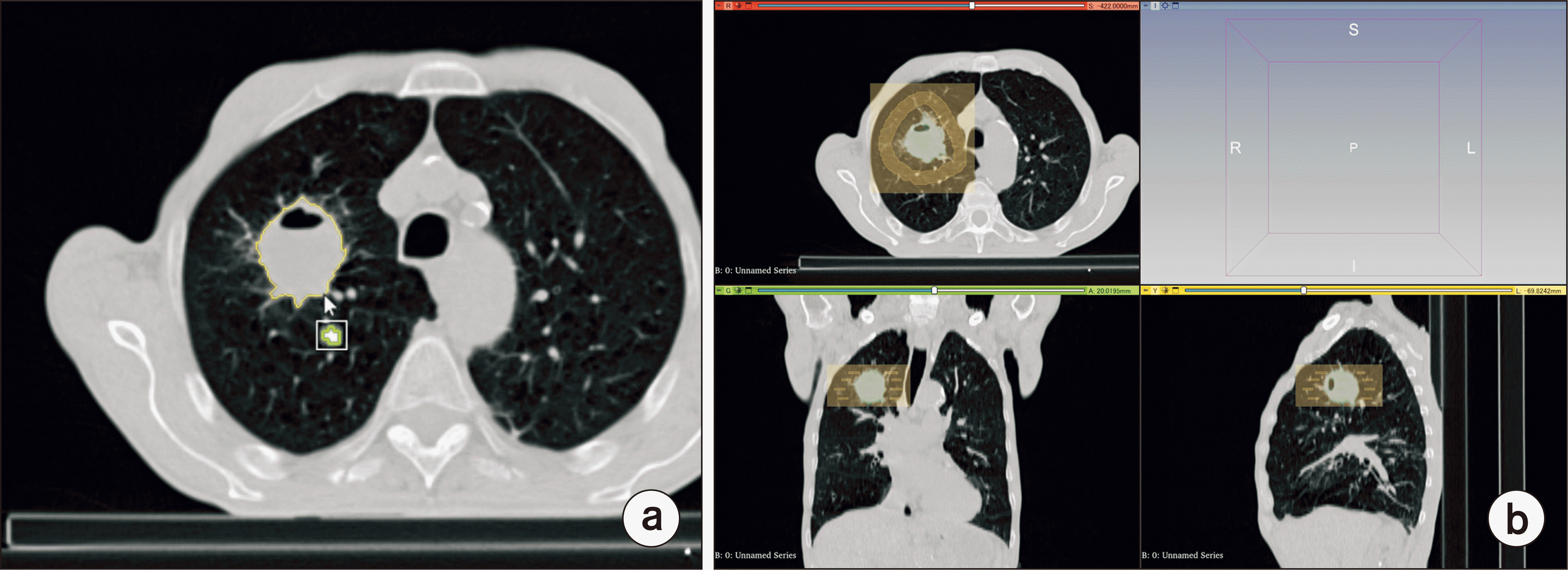
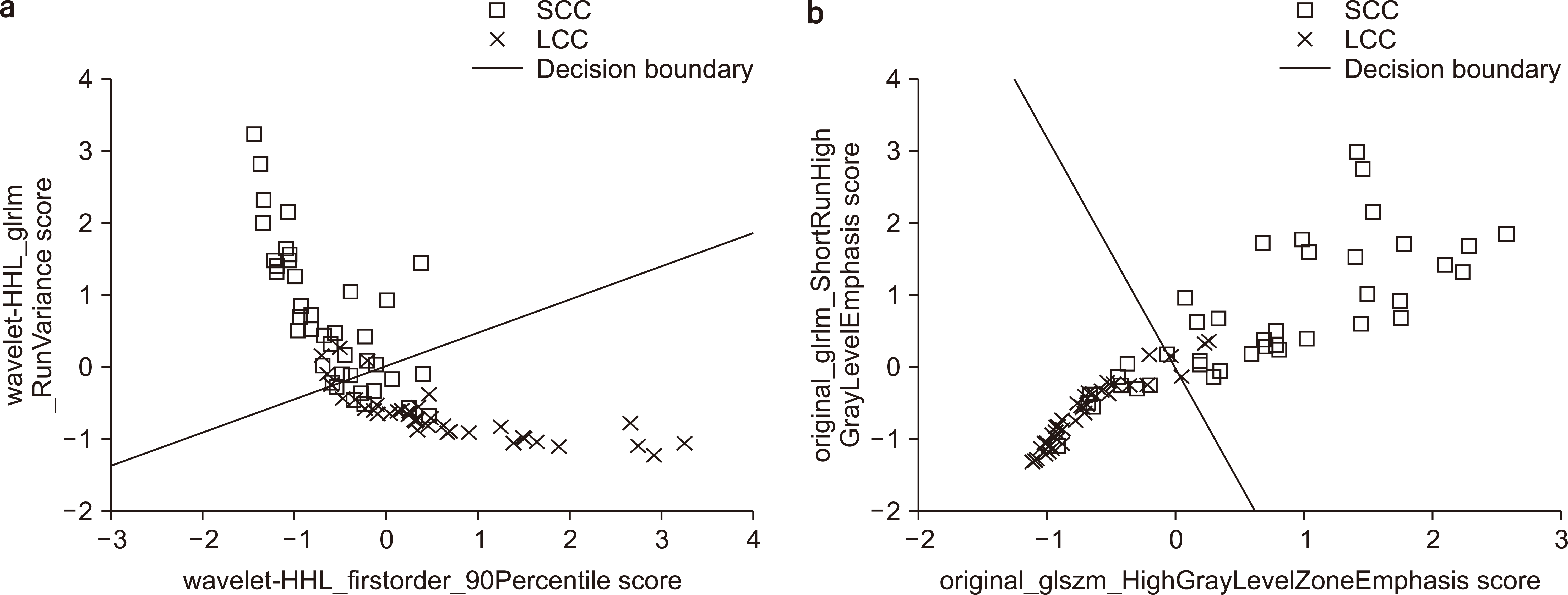
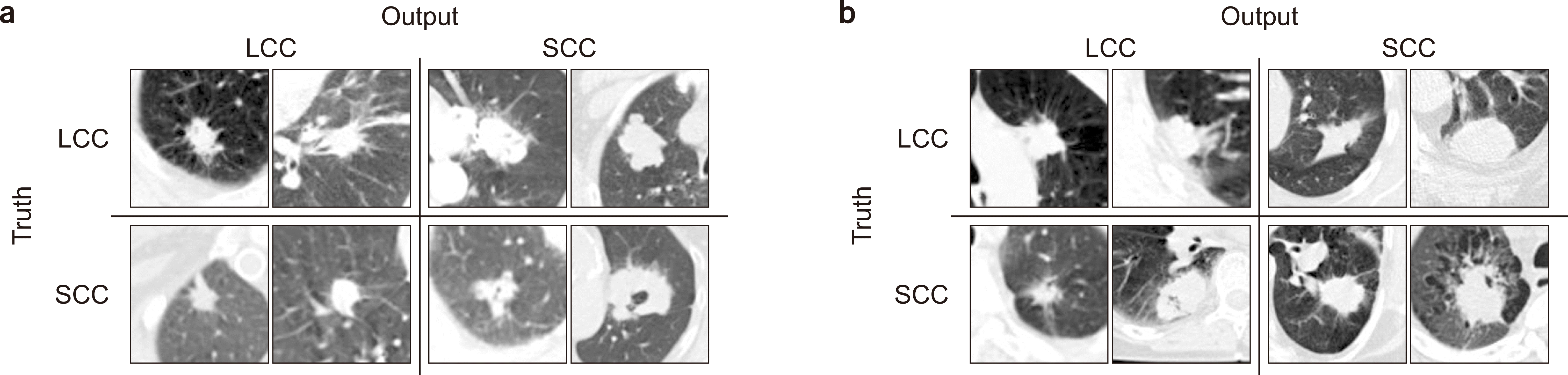
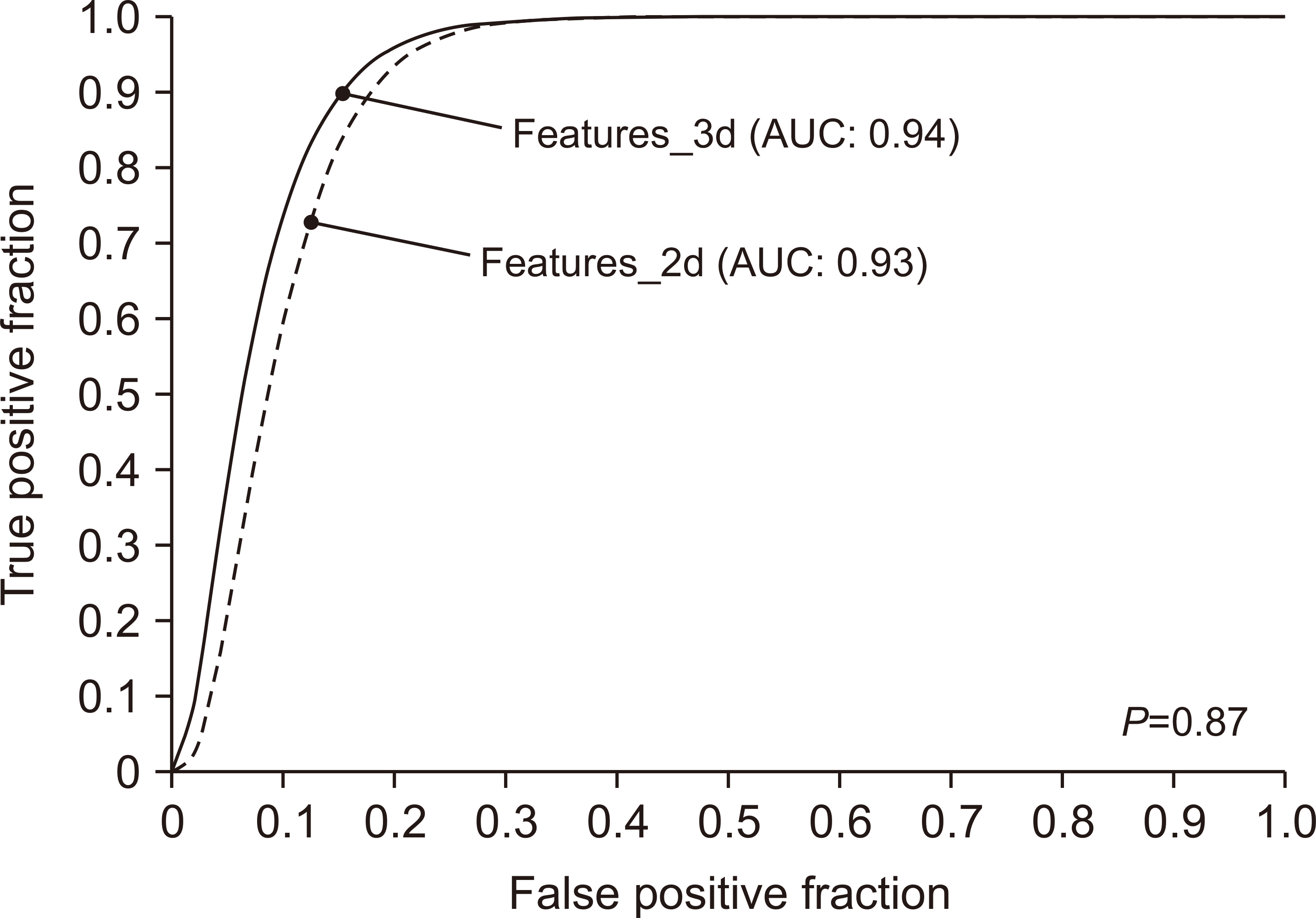




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download