Abstract
Background
The Jra antigen is a high-prevalence red blood cell (RBC) antigen. Reports on cases of fatal hemolytic disease of the fetus and newborn and acute hemolytic transfusion reactions suggest that antibodies against Jra (anti-Jra) have potential clinical significance. Identifying anti-Jra is challenging owing to a lack of commercially available antisera. We developed an alternative approach to rapidly predict the presence of anti-Jra using the TaqMan single-nucleotide polymorphism (SNP)-genotyping method.
Methods
Residual peripheral blood samples from 10 patients suspected of having the anti-Jra were collected. Two samples with confirmed Jr(a–) RBCs and anti-Jra were used to validate the TaqMan genotyping assay by comparing the genotyping results with direct sequencing. The accuracy of the assay in predicting the presence of anti-Jra was verified through crossmatching with in-house Jr(a–) O+ RBCs.
Results
The TaqMan-genotyping method was validated with two Jr(a–) RBC- and anti-Jra-confirmed samples that showed concordant Jra genotyping and direct sequencing results. Jra genotyping for the remaining samples and crossmatching the serum samples with in-house Jr(a–) O+ RBCs showed consistent results.
Conclusions
We validated a rapid, simple, accurate, and cost-effective method for predicting the presence of anti-Jra using a TaqMan-based SNP-genotyping assay. Implementing this method in routine practice in clinical laboratories will assist in solving difficult problems regarding alloantibodies to high-prevalence RBC antigens and ultimately aid in providing safe and timely transfusions and proper patient care.
The Jra antigen, which is currently the only known antigen in the JR blood group system (International Society of Blood Transfusion 032), is a high-prevalence red blood cell (RBC) antigen that is found in the blood of over 99% of the global population [1, 2]. In 2012, Zelinski et al. [3] and Saison et al. [4] revealed that the Jra antigen is encoded by the adenosine triphosphate (ATP)-binding cassette, subfamily G, member 2 (ABCG2) gene and that homozygous null alleles of ABCG2 define the Jr(a–) phenotype. The rare Jr(a–) phenotype has been found most commonly in Asian populations, primarily in Japan, although it has also been identified in various other populations, including Caucasian, Arab, and Mexican [5-11].
Antibodies against the Jra antigen (anti-Jra) are produced when individuals with Jr(a–) phenotype are exposed to Jr(a+) RBCs either by transfusion or pregnancy [12]. Anti-Jra was first reported in 1970 by Stroup and MacIlroy, who identified five individuals exhibiting blood incompatibility with all tested RBC samples except those from each other [13]. Over the past 50 yrs since the first report, case reports on anti-Jra have been published sporadically. To date, the clinical significance of anti-Jra has not been thoroughly established because of inconsistent clinical outcomes and insufficient data reporting owing to its rarity. However, anti-Jra must be considered potentially significant given previous reports on cases of fatal hemolytic disease of the fetus and newborn (HDFN) [14-16] and acute hemolytic transfusion reaction (HTR) [17].
Along with anti-Dib and anti-Rh17, anti-Jra is one of the most frequently produced alloantibodies against high-prevalence RBC antigens reported in Korea [18]. When the presence of alloantibodies against high-prevalence RBC antigens is suspected in patients based on pretransfusion testing, rapid identification of anti-Jra is needed to provide safe and timely transfusions and proper patient care. Antibodies against high-prevalence antigens (including anti-Jra, anti-Dib, and anti-Rh17) can be identified by testing selected RBCs for rare phenotypes and by typing the patient’s RBCs with antisera to high-prevalence antigens. However, clinical laboratories face limitations in serologically identifying anti-Jra owing to an inadequate supply of commercially available typing reagents, and antibody-identification testing is typically outsourced to international reference laboratories, a process that requires weeks to yield test results.
Since the genetics of the Jra antigen were discovered in 2012, the molecular bases of the JR blood group alleles have been studied extensively. As of April 2019, the molecular bases of 28 ABCG2 null alleles had been reported [19], thereby enabling predictions of the Jra phenotype based on genotyping results possible. To the best of our knowledge, although TaqMan-genotyping assays were previously used to screen donors for the Jr(a–) phenotype [20], they have not been applied and validated for predicting the presence of anti-Jra. We describe a molecular-genetics approach for utilizing a TaqMan SNP-genotyping assay that can aid in making rapid predictions of the presence of anti-Jra and can be used as a practical substitution when serological testing cannot be conducted.
From January 2017 to July 2023, we collected 10 residual peripheral blood samples during routine pretransfusion testing. These patients were suspected of potentially having anti-Jra based on pan-agglutination reactions with similar agglutination strengths across all panel cells in antibody-identification tests (including those involving enzymatic methods), and they had negative auto-control results. The Institutional Review Board of Severance Hospital in Seoul, Korea, approved this study (approval number 4-2023-0188) and waived the requirement for informed consent.
Previous findings established that the major JRnull allele in Asians is ABCG2*01N.01 (rs72552713, c.376C>T, p.Gln126*) [3, 4, 21]. According to data from the Genome Aggregation Database (v2.1.1; https://gnomad.broadinstitute.org/) [22] and the Korean Reference Genome Database [23], this JRnull allele predominates in Korea. We used the TaqMan SNP-genotyping assay for rs72552713 to perform Jra genotyping for all collected samples. The TaqMan assay for Jra genotyping was validated by comparing the genotyping results of all samples, which were serologically found to contain Jr(a–) RBCs and anti-Jra, with direct sequencing results obtained using specific primers targeting exon 4 of ABCG2. After validating the TaqMan Jra-genotyping method, we applied it to other cases where antibodies to high-prevalence RBC antigens were suspected. In addition, we conducted serum crossmatching for each sample with the in-house Jr(a–) O+ RBCs to verify the presence of anti-Jra suggested by the TaqMan Jra genotyping results.
During routine pretransfusion testing, we performed antibody screening using an Ortho BioVue System Poly Cassette with 0.8% Selectogen I, II (Ortho Clinical Diagnostics, Raritan, NJ, USA) and an LISS/Coombs ID-Card with ID-DiaCell I-II (Bio-Rad, Hercules, CA, USA) in three steps (i.e., immediate spinning, incubation at 37°C, and an indirect antiglobulin test [IAT]). In cases where antibody screening showed positive results, antibody identification was conducted using an Ortho BioVue System Poly Cassette with 0.8% Resolve Panel C Untreated and a neutral cassette with 0.8% Resolve Panel C Ficin (Ortho Clinical Diagnostics). Antibody identification was also conducted using an LISS/Coombs ID-Card with ID-DiaPanel and an NaCl, Enzyme Test, and Cold Agglutinins ID-Card with ID-DiaPanel P (Bio-Rad). IATs were performed concurrently with an autologous control during antibody identification.
Genomic DNA was extracted from samples using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Genotyping of the rs72552713 SNP (ABCG2*01N.01; c.376C>T, p.Gln126*) was performed using a predesigned TaqMan SNP-genotyping assay (Applied Biosystems, Waltham, MA, USA). The TaqMan probes included in the assay had VIC or FAM as a 5′ reporter dye and a nonfluorescent 3′ quencher. The context sequences for rs72552713 (assay ID C__98388180_20) were AATGCAAACCCACTAATACTTACTT[G(VIC)/A(FAM)]TACCACGTAACCTGAATTACATTTG, indicating that probes labeled with VIC and FAM corresponded to ABCG2*01 (allele 1; ABCG2 reference allele; c.376C) and ABCG2*01N.01 (allele 2; ABCG2 null allele; c.376C>T), respectively. A 10 µL PCR mix was prepared for each assay by mixing 2 μL (10 ng) of genomic DNA, 0.5 μL of 20× assay mix, 5 μL of TaqPath ProAmp Master Mix (Applied Biosystems), and 2.5-μL nuclease-free water. Assays were run on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: pre-reading at 60°C for 30 secs; initial denaturation/enzyme activation at 95°C for 5 mins; and 40 cycles of denaturation at 95°C for 15 secs, annealing and extension at 60°C for 90 secs, and post-reading at 60°C for 30 secs. The PCR results were analyzed using TaqMan Genotyper Software (Applied Biosystems) and visualized in allelic discrimination plots.
Direct DNA sequencing of exon 4 of ABCG2, which includes the c.376 position, was performed by Macrogen (Seoul, Korea). The forward primer sequence was 5′-AGCATGTGTTGGAGGGAAAA-3′ and the reverse primer sequence was 5′-TGTCCTTACAAGGGCCATATTC-3′. PCR amplification was performed in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad). PCR thermocycling was performed under the following conditions: initial denaturation at 95°C for 3 mins; 30 cycles of denaturation at 95°C for 30 secs, primer annealing at 54°C for 30 secs, and extension at 72°C for 30 secs; and a final extension at 72°C for 10 mins. The PCR products were purified and sequenced with a BigDye(R) Terminator v3.1 Cycle Sequencing Kit and an ABI PRISM 3730XL Analyzer (Applied Biosystems). The results were viewed using a chromatogram viewer (FinchTV 1.4.0; Geospiza Inc., Seattle, WA, USA).
Our laboratory obtained two units of Jr(a–) O+ RBCs from the Japanese Red Cross Osaka Blood Center (Osaka, Japan) in 2021 as preparation for transfusing a Jr(a–) woman during her pregnancy. The patient was discharged without receiving any transfusions, and the donated Jr(a–) O+ RBCs were cryopreserved via a modified high-glycerol method, which is typically used in clinical applications [24-28]. Briefly, the RBC concentrates were glycerolized to a final glycerol concentration of 40% using Glycerolyte 57 solution (Baxter Healthcare, Deerfield, IL, USA). An automatic dispenser (DOSE IT, Integra Biosciences, Tokyo, Japan) was then used to dispense aliquots of the glycerolized RBCs (1 mL) into several cryotubes (SPL Life Sciences, Gyeonggi-do, Korea). These cryotubes were then frozen in a controlled-rate cryo-freezer (Thermo Fisher Scientific) and stored at −70°C in an ultra-low temperature freezer (Thermo Fisher Scientific).
In cases where antibodies against high-prevalence RBC antigens were expected, one of two cryotubes containing frozen in-house Jr(a–) O+ RBCs was thawed and used in a crossmatching experiment with the patient’s serum. Cryopreserved RBCs were thawed by incubating cryotubes in a 37°C water bath for 2–3 mins with gentle mixing. The RBCs were then transferred to test tubes, treated with 1 mL of 9% NaCl, and incubated at room temperature (approximately 20–25°C) for at least 1 min. Subsequently, the RBCs were centrifuged at 3,400 rpm (approximately 1,000–1,020×g) for 20 secs, and the supernatant was removed. Subsequently, three steps (mixing 1 mL of 2.5% NaCl, centrifuging, and removing the supernatant) were repeated until a clear supernatant was observed [24]. Crossmatching was performed with the thawed RBCs using the column-agglutination technique (CAT) at 37°C during the antihuman globulin phase with the LISS/Coombs Gel Card (Bio-Rad).
Ten samples (samples 1–10, i.e., S1–S10) were included in this study (Table 1). All 10 samples were positive in antibody-screening tests and showed pan-agglutination reactions with a negative auto-control in subsequent antibody-identification tests. Enzymatic testing of the samples showed various results. Different reactions between samples and the two CAT systems were found with each sample. With S1–S8, the reaction intensities either decreased or showed a double cell population (dcp) with ficin or papain-treated RBCs. With S9 and S10, the reaction intensities were unaffected, increased, or showed a dcp with ficin or papain-treated RBCs (Supplemental Data Table S1). The presence of antibodies against high-prevalence RBC antigens, including anti-Jra, was suspected for S1–S10. Among these samples, two samples (S1 and S2) were referred to domestic or international reference laboratories, which confirmed the Jr(a–) phenotype and the presence of anti-Jra in both samples. The remaining eight samples (i.e., S3–S10) were not referred to reference laboratories and had no serological confirmation in terms of the specificity of antibodies; thus, they were considered “blind samples.”
Jra genotyping with the TaqMan SNP-genotyping assay for rs72552713 (ABCG2*01N.01; c.376C>T, p.Gln126*), and direct sequencing of ABCG2 exon 4 was performed with both samples with a confirmed Jr(a–) phenotype and anti-Jra production (S1 and S2). With both samples, the TaqMan assay demonstrated amplification of the FAM signal only. The direct sequencing results revealed homozygosity for the c.376C>T nucleotide change, indicating 100% concordance between both test methods and support for the Jr(a–) phenotype (Fig. 1). These findings validated the TaqMan SNP-genotyping method for Jra genotyping.
Eight blind samples, S3–S10, were analyzed for their Jra genotype via the validated TaqMan SNP-genotyping method. Among them, six samples (S3–S8) displayed amplification of just the FAM signal, indicating ABCG2*01N.01 homozygosity. Predictions could be made that these samples had the Jr(a–) phenotype and that anti-Jra was produced. In contrast, S9 and S10 only showed amplification of the VIC signal, which was indicative of ABCG2*01 homozygosity. Hence, these samples were predicted to have the Jr(a+) phenotype and to produce antibodies with specificities other than anti-Jra. The genotyping results are presented in the allelic discrimination plot (Fig. 2).
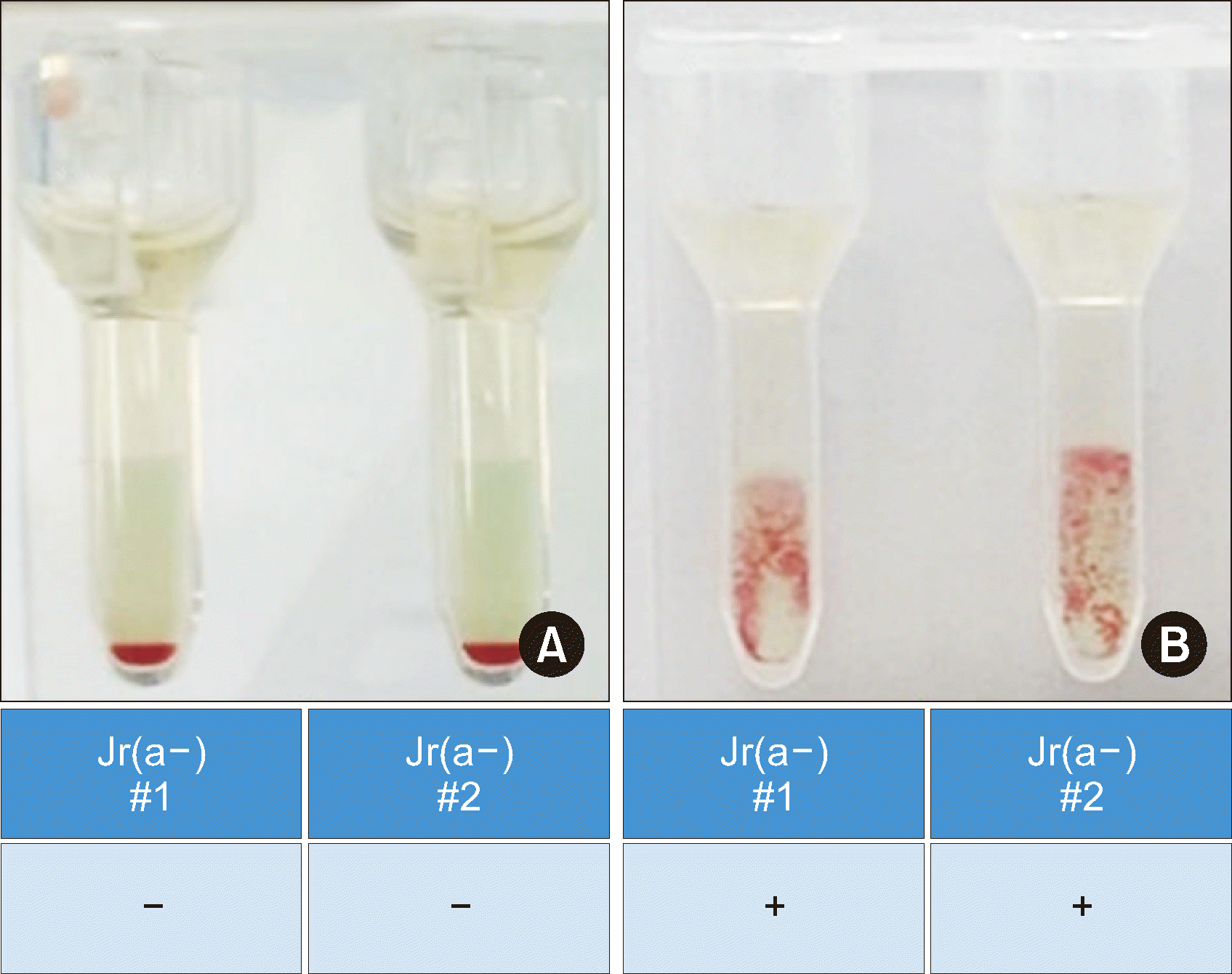
To verify the predictions regarding the presence of anti-Jra made based on the Jra-genotyping results of the TaqMan assay, we performed crossmatching between serum samples and both Jr(a–) O+ RBC samples identified in-house. The crossmatching results of S3–S8 showed no agglutination, whereas those of S9 and S10 showed agglutination (Fig. 3). Our crossmatching results were perfectly consistent with the predictions based on the TaqMan SNP-genotyping assay, demonstrating that using the TaqMan SNP-genotyping method can be a reliable strategy for predicting the presence of anti-Jra.
Cases of alloantibodies against high-prevalence antigens are rarely reported, and their clinical significance is unknown. However, anti-Jra, anti-Dib, and anti-Rh17, which are the most frequently occurring alloantibodies to high-prevalence antigens in Korea, have been associated with numerous cases of fatal HDFN or HTR and have been deemed clinically relevant [18]. Quickly identifying or excluding the presence of these potentially clinically important antibodies is important during routine pretransfusion testing for safe and timely transfusions. However, because clinical laboratories are rarely equipped with antisera against high-prevalence antigens and RBCs for these rare phenotypes, identifying these antibodies by serological testing is impractical in most cases.
To overcome this difficulty, one might think that observing the reaction patterns in enzymatic assays could help determine the specificity of these antibodies; however, the reaction patterns found with enzymatic methods should be considered carefully. For instance, the Jra antigen was reportedly resistant to ficin and papain [2]. However, our enzymatic assays showed that Jra antigen was not always resistant to ficin and papain. Agglutination decreased or showed a dcp in enzyme tests, and the Jra antigen appeared to have been destroyed or altered by the enzymes. When anti-Jra is suspected and antibody identification is performed, enzymatic data should be carefully interpreted while recognizing the possibility of inconsistent results to avoid confusion.
Accordingly, an alternative approach that can aid in anticipating the presence of anti-Jra, anti-Dib, and anti-Rh17 antibodies is required in clinical laboratories. The presence of anti-Rh17 can be predicted based on positivity for the D antigen and negativity for the C, c, E, and e antigens by performing serological Rh phenotyping [29]. However, for anti-Jra and anti-Dib, antisera are not available, and using a molecular-genetics method to determine the antigen genotype is another promising option that can help predict the existence of these antibodies. In this study, we validated and verified a rapid, simple, accurate, and affordable method for predicting anti-Jra by applying the TaqMan SNP-genotyping assay for rs72552713 (ABCG2*01N.01; c.376C>T, p.Gln126*). This method enabled discrimination between ABCG2*01 (ABCG2 reference allele) and ABCG2*01N.01 (ABCG2 null allele), and the Jra genotype was determined accurately. Comparison with direct sequencing validated the method by showing complete concordance. Predictions on whether the specificity of the alloantibody was anti-Jra, based on the Jra genotype, were confirmed by crossmatching serum samples with in-house Jr(a–) O+ RBCs. Of the 10 samples analyzed, S1–S8 were revealed to have the ABCG2*01N.01/01N.01 genotype (homozygous for the ABCG2 null allele) and to contain anti-Jra, whereas S9 and S10 were found to have the ABCG2*01/01 genotype (homozygous for the ABCG2 reference allele) and alloantibodies without specificity for anti-Jra.
One of the many advantageous features of the TaqMan SNP-genotyping method is its ability for easy multiplexing. Conducting a multiplex TaqMan SNP-genotyping assay for the Jra and Dib genotypes may be more efficient because it enables concurrent investigation for both genotypes in a single assay. Simultaneously, for samples S9 and S10, predicted to exhibit the Jr(a+) phenotype and containing alloantibodies with specificities other than anti-Jra, the multiplex TaqMan SNP-genotyping assay included a predesigned TaqMan SNP-genotyping assay for the Diego blood group targeting rs2285644, and the homozygous genotype DI*01/01 (homozygous for c.2561C>T) was detected. Thus, predictions that these samples had a Di(a+b–) phenotype and an alloantibody specificity of anti-Dib could be made.
Technologies in molecular blood group antigen (BGA) genotyping have gradually evolved. Currently, the genotypes of many BGAs can be determined simultaneously through next-generation sequencing (NGS). However, NGS remains expensive, labor-intensive, and time-consuming and requires standardization and validation before being routinely used for BGA genotyping in clinical laboratories [30]. In contrast, TaqMan SNP-genotyping assays are relatively cost-effective, simple, and fast; have been extensively validated; and have been widely applied in various routine tests in clinical laboratories. In addition, because the ABCG2*01N.01/01N.01 genotype for the Jr(a–) phenotype is overwhelmingly dominant in the Korean population, the TaqMan SNP-genotyping method can be readily adopted in most clinical laboratories in Korea to aid in predicting the presence or absence of anti-Jra, and such testing takes a few hours. However, a limitation of TaqMan SNP genotyping is the challenge of designing assays to cover all types of alleles in ethnicities with allelic heterogeneity.
This study also had several other limitations. First, TaqMan SNP genotyping could only predict the presence of Jra with a high probability but could not determine the presence of anti-Jra. When an antibody against a high-prevalence antigen is suspected based on pretransfusion tests, if the Jra genotype is confirmed [corresponding to the Jr(a–) phenotype], then it is more reasonable to consider that the antibody is specific for anti-Jra rather than another high-prevalence antigen that is also negative (which would be highly unlikely with an extremely low probability). However, the fact that the probability is extremely low does not mean that such a case cannot occur. Although TaqMan Jra genotyping can perform high-probability predictions regarding the presence of anti-Jra, a limitation is that it does not prove the presence of anti-Jra.
A second limitation of this study is that we did not include controls and samples with a heterozygous genotype (ABCG2*01/ 01N.01). The TaqMan assays were performed with three types of controls, namely a no-template control, a negative control [ABCG2*01/01 genotype, Jr(a+) phenotype], and a positive control [ABCG2*01N.01/01N.01 genotype, Jr(a–) phenotype]. Distilled water was used as no-template control. Blood donor samples showing the ABCG2*01/01 genotype in TaqMan assays, confirmed by direct sequencing of ABCG2 exon 4 (homozygous for c.376C), were included in the assays as negative controls. S1 and S2 were confirmed to have the Jr(a–) phenotype and anti-Jra by reference laboratories. TaqMan assays showed that S1 and S2 had the ABCG2*01N.01/01N.01 genotype, which was confirmed by directly sequencing ABCG2 exon 4 (homozygous for c.376C>T); we included S1 and S2 in the assays as positive controls. However, controls and samples with a heterozygous genotype (ABCG2*01/01N.01) were unavailable because such samples are difficult to obtain (as the Jra antigen is a high-prevalence RBC antigen). Although the assay must help distinguish the homozygous genotype (ABCG2*01N.01/01N.01), which indicates Jr(a–) phenotype, from the heterozygous genotype, which indicates Jr(a+) phenotype, we could not assess this aspect in this study.
A third limitation of this study is that the performance characteristics (such as precision, sensitivity, and specificity) could not be evaluated owing to difficulties in collecting a sufficient number of these rare samples. Further studies with a sufficient sample size will need to be conducted to evaluate the performance characteristics thoroughly. Finally, applying the multiplex TaqMan SNP-genotyping assay for determining the Jra and Dib genotype (to predict the presence of anti-Jra and anti-Dib) in a single assay needs to be evaluated further in future research for clinical applications.
In conclusion, we demonstrated that the TaqMan SNP-genotyping assay is an effective and practical method for Jra genotyping that can help predict the presence of anti-Jra. The multiplex TaqMan SNP-genotyping method can be implemented in routine clinical laboratories to help predict the presence or absence of troublesome anti-Jra and anti-Dib antibodies in a single assay. We plan on applying the TaqMan SNP-genotyping method in routine practice to assist in solving difficult problems regarding alloantibodies to high-prevalence RBC antigens.
Notes
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.3343/alm.2023.0325
AUTHOR CONTRIBUTIONS
Koo YK collected data and wrote the manuscript. Suh EJ, Kim NH, and Kim HK performed the experiments and collected data. Cho YK collected data and reviewed and edited the manuscript. Kwon SS and Choi SJ conceptualized, designed, and supervised the study and reviewed and edited the manuscript. Kim S and Lee KA supervised the study and reviewed the manuscript. All authors have read and approved the final manuscript.
References
1. Cohn CS, Delaney M, editors. 2020. Technical manual. 20th ed. American Association of Blood Banks;Bethesda: p. 380. DOI: 10.22215/wirl/2020.1.
2. Reid ME, Lomas-Francis C. 2012. The blood group antigen facts book. 3rd ed. Academic Press;Cambridge: p. 635–9. DOI: 10.1016/B978-0-12-415849-8.00034-X. PMID: 22258490. PMCID: PMC3677715.
3. Zelinski T, Coghlan G, Liu XQ, Reid ME. 2012; ABCG2 null alleles define the Jr(a-) blood group phenotype. Nat Genet. 44:131–2. DOI: 10.1038/ng.1075. PMID: 22246507.

4. Saison C, Helias V, Ballif BA, Peyrard T, Puy H, Miyazaki T, et al. 2012; Null alleles of ABCG2 encoding the breast cancer resistance protein define the new blood group system Junior. Nat Genet. 44:174–7. DOI: 10.1038/ng.1070. PMID: 22246505. PMCID: PMC3653631.

5. Tritchler JE. 1977; An example of anti-Jra. Transfusion. 17:177–8. DOI: 10.1046/j.1537-2995.1977.17277151926.x. PMID: 850936.
6. Kendall AG. 1976; Clinical importance of the rare erythrocyte antibody anti-Jra. Transfusion. 16:646–7. DOI: 10.1046/j.1537-2995.1976.16677060250.x. PMID: 996925.

7. Levene C, Sela R, Dvilansky A, Yermiahu T, Daniels G. 1986; The Jr(a-) phenotype and anti-Jra in two Beduin Arab women in Israel. Transfusion. 26:119–20. DOI: 10.1046/j.1537-2995.1986.26186124019.x. PMID: 3946000.

8. Vedo M, Reid ME. 1978; Anti-Jra in a Mexican American. Transfusion. 18:569. DOI: 10.1046/j.1537-2995.1978.18579036387.x. PMID: 705864.

9. Yamaguchi H, Okubo Y, Seno T, Tomita T, Ogawa Y, Fujii I, et al. 1976; A rare phenotype blood Jr(a-) occurring in two successive generations of a Japanese family. Proc Jpn Acad. 52:521–3. DOI: 10.2183/pjab1945.52.521.

10. Nakajima H, Ito K. 1978; An example of anti-Jra causing hemolytic disease of the newborn and frequency of Jra antigen in the Japanese population. Vox Sang. 35:265–7. DOI: 10.1111/j.1423-0410.1978.tb02932.x. PMID: 567888.

11. Okubo Y. 1978; Some rare blood group phenotypes in Japanese, Fy (a-), Di (b-) and Jr (a-). Blood Programme. 1:279–84. DOI: 10.5924/abgri1972.1978.1.
12. Castilho L, Reid ME. 2013; A review of the JR blood group system. Immunohematology. 29:63–8. DOI: 10.21307/immunohematology-2019-126. PMID: 24094238.

13. Stroup M, MacIlroy M. Five examples of an antibody defining an antigen of high frequency in the Caucasian population. Proceedings of the 23rd Annual Meeting of the American Association of Blood Banks. 1970; San Francisco. p. 86.
14. Peyrard T, Pham BN, Arnaud L, Fleutiaux S, Brossard Y, Guerin B, et al. 2008; Fatal hemolytic disease of the fetus and newborn associated with anti-Jr. Transfusion. 48:1906–11. DOI: 10.1111/j.1537-2995.2008.01787.x. PMID: 18522708.

15. Arriaga F, Gomez I, Linares MD, Gascon A, Carpio N, Perales A. 2009; Fatal hemolytic disease of the fetus and newborn possibly due to anti-Jr. Transfusion. 49:813. DOI: 10.1111/j.1537-2995.2008.02087.x. PMID: 19335378.

16. Kim MS, Kim JS, Park H, Chung Y, Kim H, Ko DH, et al. 2020; Fatal hemolytic disease of the fetus and newborn caused by anti-Jra antibody: a case report and literature review. Transfus Apher Sci. 59:102605. DOI: 10.1016/j.transci.2019.06.029. PMID: 31324575.
17. Kwon MY, Su L, Arndt PA, Garratty G, Blackall DP. 2004; Clinical significance of anti-Jra: report of two cases and review of the literature. Transfusion. 44:197–201. DOI: 10.1111/j.1537-2995.2004.00643.x. PMID: 14962310.

18. Choi SJ, Chung YN, Cho D, Kim S. 2019; Alloantibodies to high-incidence antigen: review of cases and transfusion experiences in Korea. Korean J Blood Transfus. 30:101–12. DOI: 10.17945/kjbt.2019.30.2.101.

19. International Society of Blood Transfusion. Names for JR (ISBT 032) blood group alleles. https://www.isbtweb.org/resource/032jr.html. Updated on July 2023.
20. Haer-Wigman L, Ait Soussan A, Ligthart P, de Haas M, van der Schoot CE. 2014; Molecular analysis of immunized Jr(a-) or Lan- patients and validation of a high-throughput genotyping assay to screen blood donors for Jr(a-) and Lan- phenotypes. Transfusion. 54:1836–46. DOI: 10.1111/trf.12544. PMID: 24456066.
21. Hue-Roye K, Lomas-Francis C, Coghlan G, Zelinski T, Reid ME. 2013; The JR blood group system (ISBT 032): molecular characterization of three new null alleles. Transfusion. 53:1575–9. DOI: 10.1111/j.1537-2995.2012.03930.x. PMID: 23066723.
22. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. 2020; The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581:434–43. DOI: 10.1038/s41586-020-2308-7. PMID: 32461654. PMCID: PMC7334197.

23. Jung KS, Hong KW, Jo HY, Choi J, Ban HJ, Cho SB, et al. 2020; KRGDB: the large-scale variant database of 1722 Koreans based on whole genome sequencing. Database (Oxford). 2020:baz146. DOI: 10.1093/database/baz146. PMID: 32133509. PMCID: PMC7056612.

24. Cho YK, Kim S, Lim J, Ko DH, Choi SJ, Kim HO. 2022; Development of cryopreserved red blood cell panels as biological reference standards for performance evaluation of ABO and D blood grouping reagents. Lab Med Online. 12:1–10. DOI: 10.47429/lmo.2022.12.1.1.

25. Song S, Choi J, Kim S, Kim HO, Min H, Kim J, et al. 2009; Development of cryopreserved red blood cell panels for verifying ABO and D blood grouping reagents. Korean J Blood Transfus. 20:46–54.
26. Meryman HT, Hornblower M. 1972; A method for freezing and washing red blood cells using a high glycerol concentration. Transfusion. 12:145–56. DOI: 10.1111/j.1537-2995.1972.tb00001.x. PMID: 5026166.

27. Högman CF, Hornblower MLS, Flodin M, Gillberg G, Meryman HT, Säfwenberg J. 1986; A simple method for high-quality frozen red cells in blood group serology. Transfusion. 26:434–6. DOI: 10.1046/j.1537-2995.1986.26587020120.x. PMID: 3765037.

28. Lelkens CCM, Noorman F, Koning JG, Truijens-de Lange R, Stekkinger PS, Bakker JC, et al. 2003; Stability after thawing of RBCs frozen with the high-and low-glycerol method. Transfusion. 43:157–64. DOI: 10.1046/j.1537-2995.2003.00293.x. PMID: 12559010.
29. Yun JW, Kang ES, Ki CS, Koh KC, Kim DW. 2012; Sensitization to multiple rh antigens by transfusion of random donor platelet concentrates in a -D- phenotype patient. Ann Lab Med. 32:429–32. DOI: 10.3343/alm.2012.32.6.429. PMID: 23130343. PMCID: PMC3486938.

30. Orzińska A. 2023; Next generation sequencing and blood group genotyping: a narrative review. Ann Blood. 8:4. DOI: 10.21037/aob-21-39.

Fig. 1
Genotyping results for S1 obtained with a TaqMan SNP assay and by direct sequencing. (A) Amplification plot of the TaqMan SNP-genotyping assay for rs72552713 (ABCG2*01N.01; c.376C>T, p.Gln126*). The increase in the fluorescent intensity of the FAM signal and not the VIC signal indicates homozygosity for c.376C>T. (B) Sequencing chromatogram obtained by directly sequencing ABCG2 exon 4. The blue arrow indicates the position showing homozygosity for c.376C>T.
Abbreviations: S1, sample 1; SNP, single-nucleotide polymorphism.
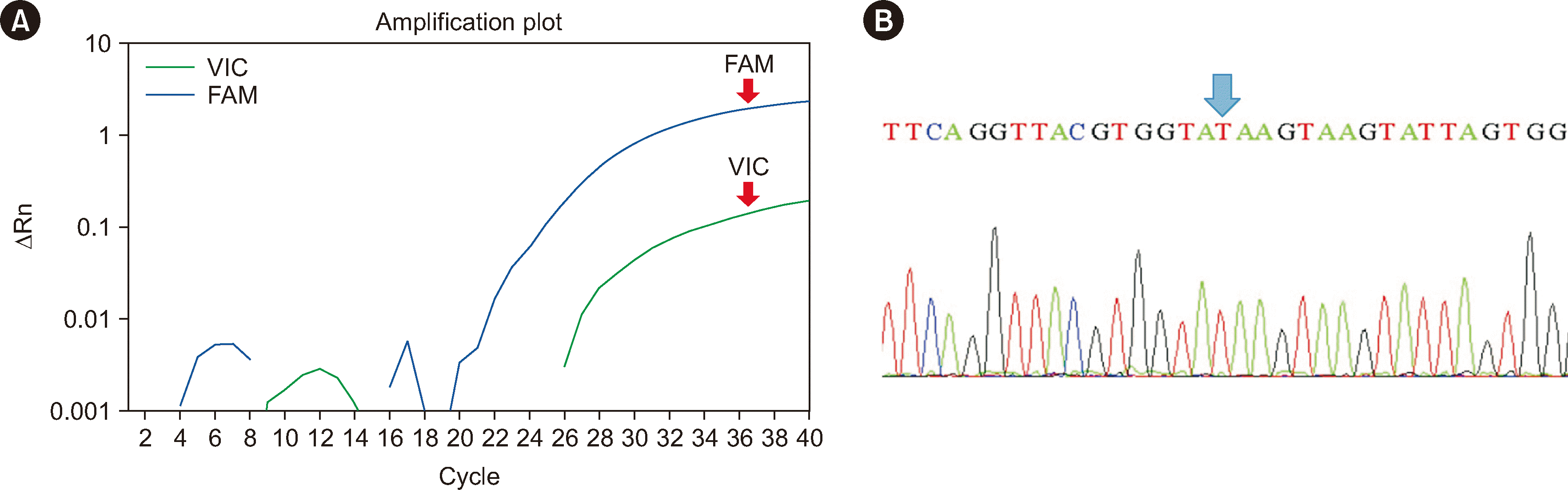
Fig. 2
TaqMan SNP-genotyping assay results visualized on an allelic discrimination plot for the ABCG2 reference allele (allele 1, VIC, ABCG2*01, c.376C) versus the ABCG2 null allele (allele 2, FAM, ABCG2*01N.01, c.376C>T). (A) The samples plotted in the upper left corner had high FAM fluorescence but low VIC fluorescence, indicating homozygosity for ABCG2*01N.01. The blue dots correspond to S1–S8. (B) The samples plotted in the lower right corner had increased VIC fluorescence but not increased FAM fluorescence, indicating homozygosity for ABCG2*01. The red dots correspond to S9 and S10.
Abbreviation: SNP, single-nucleotide polymorphism.
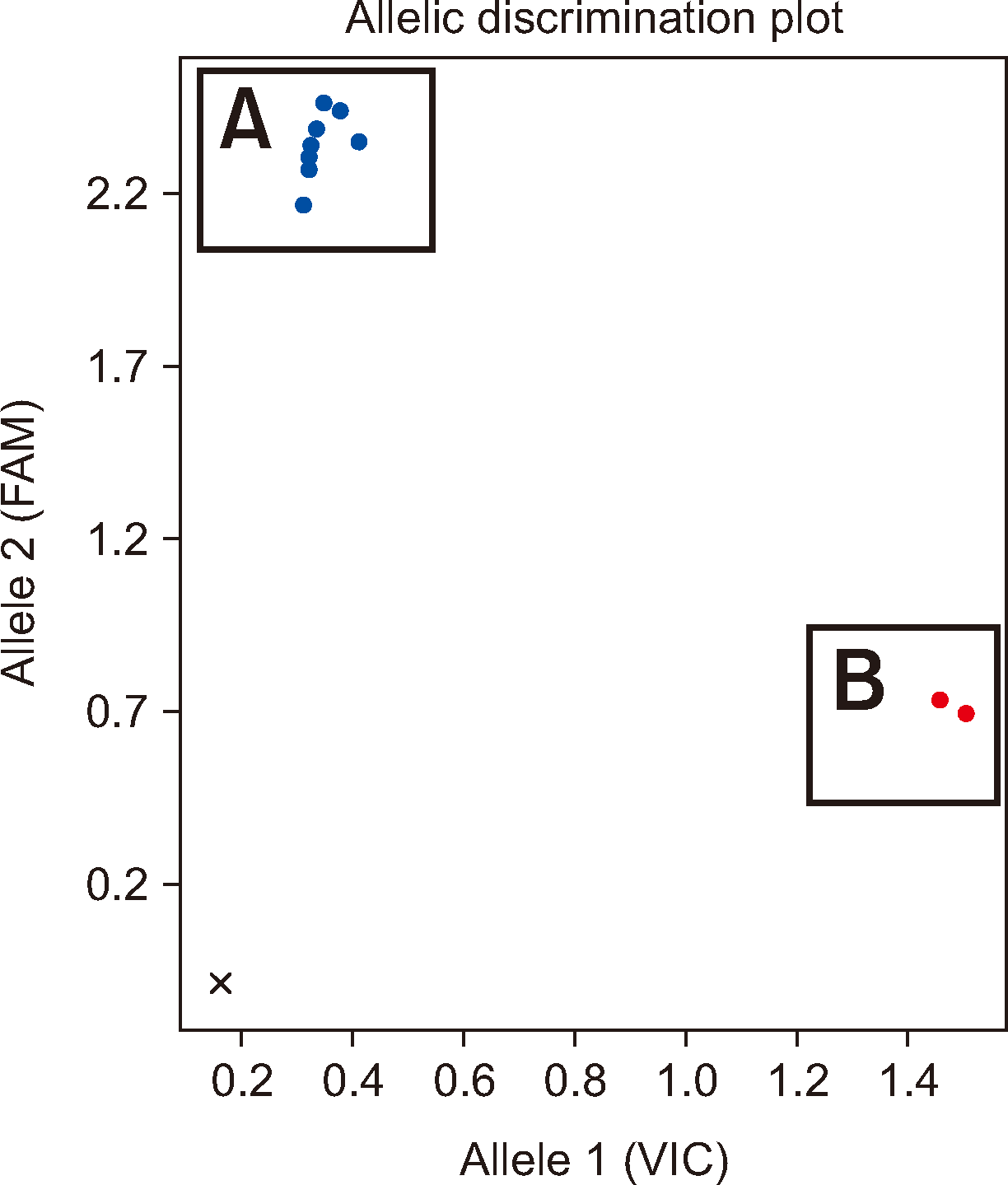
Fig. 3
Crossmatching results between two serum samples and the two in-house Jr(a–) O+ RBCs. (A) Representative crossmatching results for S6 showing no agglutination. (B) Representative crossmatching results for S9, showing agglutination.
Abbreviations: RBCs, red blood cells; S6, sample 6.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download