Abstract
Background
Carbapenem resistance in Pseudomonas aeruginosa is a serious global health problem. We investigated the clonal distribution and its association with the carbapenem resistance mechanisms of carbapenem-non-susceptible P. aeruginosa isolates from three Korean hospitals.
Methods
A total of 155 carbapenem-non-susceptible P. aeruginosa isolates collected between 2011 and 2019 were analyzed for sequence types (STs), antimicrobial susceptibility, and carbapenem resistance mechanisms, including carbapenemase production, the presence of resistance genes, OprD mutations, and the hyperproduction of AmpC β-lactamase.
Results
Sixty STs were identified in carbapenem-non-susceptible P. aeruginosa isolates. Two high-risk clones, ST235 (N=41) and ST111 (N=20), were predominant; however, sporadic STs were more prevalent than high-risk clones. The resistance rate to amikacin was the lowest (49.7%), whereas that to piperacillin was the highest (92.3%). Of the 155 carbapenem-non-susceptible isolates, 43 (27.7%) produced carbapenemases. Three metallo-β-lactamase (MBL) genes, blaIMP-6 (N=38), blaVIM-2 (N=3), and blaNDM-1 (N=2), were detected. blaIMP-6 was detected in clonal complex 235 isolates. Two ST773 isolates carried blaNDM-1 and rmtB. Frameshift mutations in oprD were identified in all isolates tested, regardless of the presence of MBL genes. Hyperproduction of AmpC was detected in MBL gene–negative isolates.
Pseudomonas aeruginosa is an important nosocomial pathogen that causes various opportunistic infections, including pneumonia and urinary tract, bloodstream, and surgical wound infections, primarily in immunocompromised or severely ill patients [1, 2]. This microorganism is notorious for its ability to develop antimicrobial resistance [3, 4]. Multidrug-resistant (MDR) or extensively drug-resistant (XDR) P. aeruginosa strains significantly increase morbidity and mortality rates in infected patients [4-6]. The WHO has listed carbapenem-resistant P. aeruginosa (CRPA) as a critical priority pathogen for the development of new antimicrobials [7].
Carbapenems are commonly used to treat MDR P. aeruginosa infections, which has resulted in the prevalence of CRPA [4]. Carbapenem resistance in P. aeruginosa is attributed to intrinsic resistance mechanisms, including reduced membrane permeability, overexpression of efflux pumps, or hyperproduction of de-repressed AmpC β-lactamase, or the acquisition of diverse genes encoding carbapenem-hydrolyzing enzymes [8-12]. Ambler class B metallo-β-lactamases (MBLs) confer a high level of resistance to carbapenems and are highly associated with global epidemic clones [6]. Hyperproduction of chromosomal AmpC β-lactamases is another important mechanism of resistance to cephalosporins and carbapenems [13]. Inducible AmpC by certain β-lactams and mutations in the regulatory components of AmpC leads to stable hyperproduction of AmpC. The outer membrane porin OprD allows the entrance of carbapenems into the periplasmic space in P. aeruginosa [14, 15]. Loss or alteration of OprD by mutation leads to carbapenem resistance.
The dissemination of global epidemic clones is responsible for the worldwide prevalence of MDR P. aeruginosa isolates. Based on prevalence, global spread, association with MDR and XDR profiles, and horizontally acquired β-lactamases, such as extended-spectrum β-lactamases and carbapenemases, del Barrio-Tofiño et al. [6] suggested top 10 P. aeruginosa high-risk clones globally, including sequence type (ST) 111, ST175, ST233, ST235, ST244, ST277, ST298, ST308, ST357, and ST654. Among them, ST235 was the most widespread and carried over 60 different β-lactamase gene variants [16, 17]. The prevalence of ST235 P. aeruginosa carrying blaIMP-6 or blaVIM-2 in Korea has been reported [18-20]. We investigated the clonal distribution and its association with carbapenem resistance mechanisms of carbapenem-non-susceptible P. aeruginosa isolates from three hospitals located in southern Korea to obtain a detailed perspective on the molecular epidemiology of carbapenem resistance in P. aeruginosa. This study demonstrated that diverse clones of P. aeruginosa, including global high-risk clones, are circulating in Korean hospitals. Carbapenem resistance in Korean P. aeruginosa isolates is mediated by frameshift mutations in oprD combined with MBL production or ampC overexpression.
A total of 155 carbapenem-non-susceptible P. aeruginosa isolates from three Korean hospitals, Kyungpook National University Hospital (KNUH) located in Daegu (N=50), Gyeongsang National University Hospital (GNUH) located in Jinju (N=45), and Jeonbuk National University Hospital (JNUH) located in Jeonju (N=60), collected between 2011 and 2019, were used in this study (Supplemental Data Table S1). The isolates were obtained from three regional branches of the National Culture Collection for Pathogens (NCCP), KNUH-NCCP, GNUH-NCCP, and JNUH-NCCP, which collect and deposit representative bacterial pathogen resources from associated hospitals. Of the 345 carbapenem-non-susceptible P. aeruginosa isolates deposited in KNUH-NCCP (N=111), GNUH-NCCP (N=89), and JNUH-NCCP (N=145), 155 (44.9%) isolates were randomly selected based on the isolation year and date, source clinical samples, antimicrobial susceptibility profiles, and patient clinical information. The isolates were obtained in 2011 (N=10 of 24 isolates), 2012 (N=24 of 54), 2013 (N=28 of 59), 2014 (N=8 of 19), 2015 (N=14 of 27), 2016 (N=10 of 25), 2017 (N=21 of 47), 2018 (N=24 of 51), and 2019 (N=16 of 39). Carbapenem-non-susceptible P. aeruginosa isolates were obtained from blood (N=50 of 101 isolates), sputum and respiratory tract (N=43 of 99), urine (N=28 of 61), ascites and bile juice (N=14 of 31), wound and pus (N=13 of 30), cerebrospinal fluid and other body fluids (N=3 of 10), and other samples (N=4 of 13). P. aeruginosa was identified using the VITEK 2 system (bioMérieux, Marcy-l’Étoile, France), matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker Daltonics, Bremen, Germany), and 16S rDNA gene sequencing (Macrogen, Seoul, Korea). This study was approved by the Institutional Review Board of KNUH, Daegu, Korea (IRB No. 2018-11-023-005).
The antimicrobial susceptibility of the P. aeruginosa isolates was determined using the VITEK 2 system with an AST-N225 card (bioMérieux), according to CLSI guidelines [21]. Twelve antimicrobial agents were tested: aminoglycosides (amikacin, gentamicin, and tobramycin), antipseudomonal penicillins (piperacillin), antipseudomonal penicillins/β-lactamase inhibitors (piperacillin/tazobactam), antipseudomonal cephalosporins (cefepime and ceftazidime), carbapenems (imipenem and meropenem), monobactam (aztreonam), fluoroquinolones (ciprofloxacin), and polymyxins (colistin). The broth microdilution method was used to determine the minimum inhibitory concentrations (MICs) of three antimicrobial agents, including imipenem, meropenem, and colistin. Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as quality control strains. MDR or XDR phenotypes were determined according to the criteria suggested by Magiorakos et al. [22].
The Rapidec Carba NP test (bioMérieux) was used to screen for carbapenemase production in imipenem- or meropenem-non-susceptible P. aeruginosa isolates [23]. PCR and subsequent sequencing were used to detect antimicrobial resistance genes and oprD mutations. Briefly, bacteria were grown in lysogeny broth (LB) overnight, washed with phosphate-buffered saline, and resuspended in sterilized distilled water. The bacteria were boiled for 10 min to prepare template DNA for PCR. Carbapenemase-producing isolates were found to carry carbapenemase genes such as blaGES, blaKPC, blaIMP, blaNDM, blaSPM, blaVIM, blaOXA-48, and blaOXA-58 [15, 24-26]. Aminoglycoside-resistant isolates amplified 16S rRNA methyltransferase genes armA and rmtB [27]. Full-length blaIMP, blaNDM, and blaVIM were amplified by PCR and sequenced to subtype the genes [28, 29]. Full-length oprD was amplified and sequenced to identify mutations [24]. oprD nucleotide sequences were compared with those in P. aeruginosa PAO1 (accession No. NC_002516.2) using the Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST). The primers used for PCR and sequencing are listed in Supplemental Data Table S2.
Bacteria were grown in LB at 37°C under shaking overnight and then diluted to 1:100 in fresh LB. Bacterial cultures were grown until the optical density at 600 nm reached 1.0. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Random hexamer primers and TOPscript reverse transcriptase (Enzynomics, Daejeon, Korea) were used to synthesize cDNA. qPCRs were run using TOPreal qPCR 2X PreMIX (SYBR Green with high ROX) (Enzynomics) in a StepOnePlus Real-time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturers’ instructions. ampC expression levels were normalized to the levels of the housekeeping gene, rpsL [15, 30], and fold changes were calculated using the 2−ΔΔCt method [31]. Gene expression levels in clinical isolates were compared with those in P. aeruginosa PAO1. The specific primers for ampC and rpsL are listed in Supplemental Data Table S2. The assay was performed three times independently.
Seven genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were PCR-amplified and sequenced to determine the STs. The allelic numbers of each gene and STs were determined using a multilocus sequence typing database for P. aeruginosa (https://pubmlst.org/organisms/pseudomonas-aeruginosa).
A total of 155 carbapenem-non-susceptible P. aeruginosa isolates were classified into 60 STs, including eight unassigned STs (Supplemental Data Table S1). Three STs were prevalent, ST235 (N=41), ST111 (N=20), and ST245 (N=10), accounting for 45.8% of the isolates. ST111 and ST235 isolates were detected in the three hospitals, whereas ST245 isolates were detected only in one hospital (JNUH) (Fig. 1A). ST111 isolates were detected throughout the study period, whereas ST245 isolates were detected only between 2011 and 2013 (Fig. 1B). Eight carbapenem-non-susceptible P. aeruginosa isolates belonging to unassigned STs were detected after 2017. Of the three single locus variants of ST235, ST622, ST660, and ST1015, ST622 (N=6) and ST1015 (N=4) isolates were detected between 2018 and 2019.
MDR or XDR phenotypes were present in 20 (12.9%) and 124 (80.0%) of the 155 carbapenem-non-susceptible P. aeruginosa isolates, respectively (Supplemental Data Table S1). Carbapenem-non-susceptible P. aeruginosa isolates exhibited the highest resistance rate to piperacillin (92.3%), and the lowest to amikacin (49.7%) (Table 1). Resistance rates to amikacin, gentamicin, tobramycin, piperacillin, piperacillin/tazobactam, cefepime, and ciprofloxacin were higher in ST235 than in other STs. Colistin resistance was not observed. Resistance rates to the tested antimicrobial agents were similar among the hospitals (Table 2).
The Rapidec Carba NP test revealed that 43 (27.7%) of the 155 carbapenem-non-susceptible P. aeruginosa isolates produced carbapenemases (Supplemental Data Table S1). Sequencing of PCR amplicons revealed that blaIMP-6, blaVIM-2, and blaNDM-1 were present in 38, 3, and 2 isolates, respectively. All blaIMP-6 genes were detected in clonal complex (CC) 235 isolates, including ST235 (N=35), ST622 (N=1), ST660 (N=1), and ST1015 (N=1). blaVIM-2 was detected in ST235 (N=2) and ST463 (N=1) isolates. blaNDM-1 was detected in ST773 isolates. The blaIMP-6- and blaNDM-1-carrying P. aeruginosa isolates exhibited high-level resistance to meropenem (>256 µg/mL). Seventy isolates were non-susceptible to the three aminoglycosides tested. Among them, two ST773 isolates carried the 16S rRNA methyltransferase gene, rmtB.
Nineteen CRPA isolates were selected on the basis of their STs to determine whether oprD mutations and AmpC hyperproduction were responsible for their resistance to carbapenems. All isolates tested carried frameshift mutations in oprD, resulting in the formation of a premature stop codon (Table 3). In contrast to the 443 amino acids found in OprD of P. aeruginosa PAO1, the predicted number of amino acids of OprD in the isolates tested was 0–441. Hyperproduction of AmpC (i.e., ≥10-fold higher expression than in P. aeruginosa PAO1) was detected in six isolates belonging to ST244 (N=1), ST245 (N=3), ST622 (N=1), and ST1154 (N=1). The AmpC-hyperproducing isolates did not carry MBL genes.
High-risk P. aeruginosa clones are characterized by global spread, MDR/XDR phenotypes, and association with the production of extended-spectrum β-lactamases and carbapenemases [6]. ST111 (N=20), ST235 (N=41), ST244 (N=6), ST357 (N=3), and ST654 (N=1) were identified in this study, accounting for 32.9% of the isolates of the 10 high-risk clones. The wide spread of ST111, ST235, and ST244 in Korean hospitals, including the study sites, has been previously reported [18, 19, 32, 33]. ST245 was detected in Korea between 2006 and 2012 [32, 34] but not after 2014. After 2018, single locus variants of ST235, including aroE (ST622) and nuoD (ST1015) variants, were more prevalent than ST235. After 2017, eight unassigned STs that carried all aroE variations in the existing STs emerged (Supplemental Data Table S1). Our findings indicate that temporospatial clonal changes, primarily because of aroE variations, occur in Korean carbapenem-non-susceptible P. aeruginosa isolates.
CC235 P. aeruginosa isolates are notorious for being highly resistant to antimicrobial agents, including carbapenems [6]. In comparison to isolates of other STs, ST235 isolates showed higher resistance rates to aminoglycosides, piperacillin, piperacillin/tazobactam, cefepime, and ciprofloxacin. The therapeutic options for treating P. aeruginosa infections are limited by the prevalence of the carbapenem-resistant ST235 clone.
Carbapenem resistance in P. aeruginosa has been rapidly increasing via the clonal spread of high-risk clones carrying carbapenemase genes. In P. aeruginosa high-risk clones, two MBL genes, blaIMP and blaVIM, are widely distributed [6, 35]. The most frequent carbapenemase gene detected in Korean P. aeruginosa isolates was blaIMP-6, followed by blaVIM-2 [33, 36]. The presence of blaIMP-1, blaIMP-6, and blaVIM-2 in Korean ST235 isolates has been reported [18, 20]. In Korean ST298 and ST357 isolates, blaVIM-2 was also detected [18, 33]. All blaNDM-1 genes have been identified in ST773 isolates from Korean hospitals [20, 37, 38]. In recent years, blaNDM-1-carrying ST773 isolates were increasingly prevalent in Korea, mostly in hospitals located in Daejeon city and Gyeongsang-do [20, 37]. Interestingly, most blaNDM-1-carrying ST773 isolates originated from urine samples [20, 38]. In this study, two blaNDM-1- and rmtB-carrying ST773 P. aeruginosa isolates were identified in sputum and blood samples collected at GNUH, located in Gyeongsang-do. The blaIMP-6-carrying CC235 isolates from Korean hospitals harbored identical or similar class 1 integrons carrying blaIMP-6 as a gene cassette [18, 36, 39], indicating clonal spread rather than horizontal gene transfer. Of the three blaVIM-2-carrying isolates identified in this study, two were ST235 and one was ST463. blaVIM-2 can also be inserted as a gene cassette in class 1 integrons [36]. As ST463 is not clonally related to ST235 and ST357, it is possible that the ST463 isolate acquired blaVIM-2 via horizontal gene transfer. This should be confirmed by genetic analysis surrounding blaVIM-2. Our results demonstrated that the prevalence of MBL-producing P. aeruginosa in Korea is caused by the clonal spread of CC235 carrying blaIMP-6, and the emergence and spread of new clones carrying blaVIM-2 or blaNDM-1 should be carefully monitored.
Of the 155 carbapenem-non-susceptible isolates, 43 produced MBLs, whereas the remaining 112 isolates may possess other mechanisms of carbapenem resistance, such as membrane permeability alterations, efflux pump overexpression, or AmpC β-lactamase de-repression. Interestingly, all CRPA isolates tested carried frameshift mutations in oprD, resulting in no or truncated protein production. A frameshift or substitutional mutation in oprD resulting in no or very low production in the outer membrane was found in 21 of 22 carbapenem-resistant bloodstream P. aeruginosa isolates from KNUH, in line with previous study results [19]. Overexpression of efflux pump genes, including mexB for MexAB-OprM, mexD for MexCD-OprJ, mexF for MexEF-OprN, and mexY for MexXY-OprM, was not observed in a previous study [19], indicating that carbapenem resistance is not associated with overexpression of efflux pumps. Overexpression of ampC was observed in one ST308 isolate [19]. In this study, six of 19 isolates overexpressed ampC. AmpC hyperproduction was observed in MBL gene–negative isolates. It is generally responsible for resistance to cephalosporins and carbapenems [13], but one AmpC-hyperproducing ST245 isolate was susceptible to cephalosporins. Low membrane permeability is more closely associated with imipenem resistance than with meropenem resistance, whereas overexpression of efflux pumps is involved in reduced susceptibility to meropenem [15]. We previously demonstrated that blaIMP-6-carrying P. aeruginosa isolates exhibited higher meropenem MICs than imipenem MICs [19]. In this study, nine of the 13 carbapenem-resistant isolates without MBL genes showed higher MICs for imipenem than for meropenem. All P. aeruginosa isolates carrying blaIMP-6 exhibited higher meropenem MICs than imipenem MICs. Two blaNDM-1-carrying P. aeruginosa isolates exhibited high-level resistance to imipenem and meropenem (≥256 µg/mL). In light of earlier findings, the present study suggests that a combination of frameshift mutations in oprD and the production of MBLs or hyperproduction of AmpC is responsible for carbapenem resistance in Korean P. aeruginosa isolates.
One limitation of this study is the small sample size. Although we carefully selected 155 from 345 carbapenem-non-susceptible P. aeruginosa isolates collected in three NCCPs, the limited sample size may have influenced the clonal distribution of P. aeruginosa isolates. Another limitation is that recent isolates were not included; therefore, our results cannot reflect the clonal changes and carbapenem resistance mechanisms of circulating P. aeruginosa isolates from Korean hospitals. The intrinsic carbapenem resistance mechanisms of P. aeruginosa were investigated in 13 isolates selected from the 112 MBL-negative isolates, which may have resulted in underestimation of the intrinsic carbapenem resistance mechanisms. However, the results obtained in this study, including oprD mutations and ampC overexpression, were in line with those from a previous study [19]. This emphasizes the importance of intrinsic carbapenem resistance mechanisms in Korean P. aeruginosa isolates.
In conclusion, numerous clonally diverse carbapenem-non-susceptible P. aeruginosa strains are circulating in Korean hospitals. The most common high-risk P. aeruginosa clone during the study period was ST235; however, single locus variants of ST235 caused by aroE or nuoD variations have recently emerged. Carbapenem resistance in Korean P. aeruginosa isolates is caused by low drug permeability because of the mutational inactivation of OprD combined with MBL production or AmpC hyperproduction. The spread of MBL-producing clones, particularly the blaNDM-1-carrying ST773, and clonal changes in P. aeruginosa isolates should be carefully monitored.
Notes
SUPPLEMENTARY MATERIALS
Supplementary materials can be found via https://doi.org/10.3343/alm.2023.0369
AUTHOR CONTRIBUTIONS
Kim YK and Lee JC conceptualized the study. Kwon KT, Kim YK, and Lee JC contributed to data curation. Kim N, Ko SY, Park SY, Kim SY, and Lee DE contributed to methodology. Kwon KT acquired funding. Kim N and Lee JC wrote the original manuscript draft. Kim N, Kim YK, and Lee JC edited and reviewed the manuscript. All authors have read and approved the final manuscript.
References
1. Nguyen L, Garcia J, Gruenberg K, MacDougall C. 2018; Multidrug-resistant Pseudomonas infections: hard to treat, but hope on the horizon? Curr Infect Dis Rep. 20:23. DOI: 10.1007/s11908-018-0629-6. PMID: 29876674.
2. Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. 2019; Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY Antimicrobial Surveillance Program, 1997-2016. Open Forum Infect Dis. 6:S63–8. DOI: 10.1093/ofid/ofy343. PMID: 30895216. PMCID: PMC6419917.
3. Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. 2009; Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 48:1–12. DOI: 10.1086/595011. PMID: 19035777.

4. Botelho J, Grosso F, Peixe L. 2019; Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist Updat. 44:100640. DOI: 10.1016/j.drup.2019.07.002. PMID: 31492517.
5. Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, et al. 2019; Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 32:e00031–19. DOI: 10.1128/CMR.00031-19. PMID: 31462403. PMCID: PMC6730496.
6. del Barrio-Tofiño E, López-Causapé C, Oliver A. 2020; Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 56:106196. DOI: 10.1016/j.ijantimicag.2020.106196. PMID: 33045347.
7. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. 2018; Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 18:318–27. DOI: 10.1016/S1473-3099(17)30753-3. PMID: 29276051.
8. Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011; Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–26. DOI: 10.1016/j.tim.2011.04.005. PMID: 21664819.
9. Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. 2012; Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol. 302:63–8. DOI: 10.1016/j.ijmm.2011.10.001. PMID: 22226846. PMCID: PMC3831278.
10. Diene SM, Rolain JM. 2014; Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 20:831–8. DOI: 10.1111/1469-0691.12655. PMID: 24766097.
11. Botelho J, Roberts AP, León-Sampedro R, Grosso F, Peixe L. 2018; Carbapenemases on the move: it's good to be on ICEs. Mob DNA. 9:37. DOI: 10.1186/s13100-018-0141-4. PMID: 30574213. PMCID: PMC6299553. PMID: b77ce64c60be43be83e4928e9cc56da2.

12. Partridge SR, Kwong SM, Firth N, Jensen SO. 2018; Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 31:e00088–17. DOI: 10.1128/CMR.00088-17. PMID: 30068738. PMCID: PMC6148190.

13. Lister PD, Wolter DJ, Hanson ND. 2009; Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 22:582–610. DOI: 10.1128/CMR.00040-09. PMID: 19822890. PMCID: PMC2772362.

14. Farra A, Islam S, Stralfors A, Sorberg M, Wretlind B. 2008; Role of outer membrane protein OprD and penicillin-binding proteins in resistance of Pseudomonas aeruginosa to imipenem and meropenem. Int J Antimicrob Agents. 31:427–33. DOI: 10.1016/j.ijantimicag.2007.12.016. PMID: 18375104.
15. Rodríguez-Martínez JM, Poirel L, Nordmann P. 2009; Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 53:4783–8. DOI: 10.1128/AAC.00574-09. PMID: 19738025. PMCID: PMC2772299.
16. Oliver A, Mulet X, López-Causapé C, Juan C. 2015; The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 21-22:41–59. DOI: 10.1016/j.drup.2015.08.002. PMID: 26304792.
17. Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, et al. 2018; Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 24:258–66. DOI: 10.1016/j.cmi.2017.06.018. PMID: 28648860.
18. Cho HH, Kwon GC, Kim S, Koo SH. 2015; Distribution of Pseudomonas-derived cephalosporinase and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from Korea. J Microbiol Biotechnol. 25:1154–62. DOI: 10.4014/jmb.1503.03065. PMID: 25907063.
19. Kim CH, Kang HY, Kim BR, Jeon H, Lee YC, Lee SH, et al. 2016; Mutational inactivation of OprD in carbapenem-resistant Pseudomonas aeruginosa isolates from Korean hospitals. J Microbiol. 54:44–9. DOI: 10.1007/s12275-016-5562-5. PMID: 26727901.
20. Park Y, Koo SH. 2022; Epidemiology, molecular characteristics, and virulence factors of carbapenem-resistant Pseudomonas aeruginosa isolated from patients with urinary tract infections. Infect Drug Resist. 15:141–51. DOI: 10.2147/IDR.S346313. PMID: 35058697. PMCID: PMC8765443.
21. CLSI. 2020. Performance standards for antimicrobial susceptibility testing. 30th ed. Clinical and Laboratory Standards Institute;Wayne, PA: CLSI M100. DOI: 10.1016/s0196-4399(01)88009-0.
22. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. 2012; Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 18:268–81. DOI: 10.1111/j.1469-0691.2011.03570.x. PMID: 21793988.

23. Poirel L, Nordmann P. 2015; Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol. 53:3003–8. DOI: 10.1128/JCM.00977-15. PMID: 26085619. PMCID: PMC4540946.

24. Fournier D, Richardot C, Müller E, Robert-Nicoud M, Llanes C, Plésiat P, et al. 2013; Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J Antimicrob Chemother. 68:1772–80. DOI: 10.1093/jac/dkt098. PMID: 23587654.
25. Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. 2007; Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 45:544–7. DOI: 10.1128/JCM.01728-06. PMID: 17093019. PMCID: PMC1829038.

26. Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. 2012; Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 67:906–9. DOI: 10.1093/jac/dkr563. PMID: 22232516.

27. Asghar A, Ahmed O. 2018; Prevalence of aminoglycoside resistance genes in Pseudomonas aeruginosa isolated from a tertiary care hospital in Makkah, KSA. Clin Pract. 15:541–7. DOI: 10.4172/clinical-practice.1000391.
28. Kawahara R, Watahiki M, Matsumoto Y, Uchida K, Noda M, Masuda K, et al. 2021; Subtype screening of blaIMP genes using bipartite primers for DNA sequencing. Jpn J Infect Dis. 74:592–9. DOI: 10.7883/yoken.JJID.2020.926. PMID: 33790070.

29. Fiett J, Baraniak A, Mrówka A, Fleischer M, Drulis-Kawa Z, Naumiuk L, et al. 2006; Molecular epidemiology of acquired-metallo-β-lactamase-producing bacteria in Poland. Antimicrob Agents Chemother. 50:880–6. DOI: 10.1128/AAC.50.3.880-886.2006. PMID: 16495246. PMCID: PMC1426447.

30. Juan C, Moyá B, Pérez JL, Oliver A. 2006; Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob Agents Chemother. 50:1780–7. DOI: 10.1128/AAC.50.5.1780-1787.2006. PMID: 16641450. PMCID: PMC1472203.

31. Livak KJ, Schmittgen TD. 2001; Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 25:402–8. DOI: 10.1006/meth.2001.1262. PMID: 11846609.

32. Cho HH, Kwon KC, Kim S, Koo SH. 2014; Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa. Ann Lab Med. 34:286–92. DOI: 10.3343/alm.2014.34.4.286. PMID: 24982833. PMCID: PMC4071185.

33. Hong JS, Kim JO, Lee H, Bae IK, Jeong SH, Lee K. 2015; Characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother. 47:33–40. DOI: 10.3947/ic.2015.47.1.33. PMID: 25844261. PMCID: PMC4384452.

34. Lee JY, Peck KR, Ko KS. 2013; Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol. 62:1015–24. DOI: 10.1099/jmm.0.055426-0. PMID: 23558139.
35. Hammoudi Halat D, Ayoub Moubareck C. 2022; The Intriguing carbapenemases of Pseudomonas aeruginosa: current status, genetic profile, and global epidemiology. Yale J Biol Med. 95:507–15.
36. Bae IK, Suh B, Jeong SH, Wang KK, Kim YR, Yong D, et al. 2014; Molecular epidemiology of Pseudomonas aeruginosa clinical isolates from Korea producing β-lactamases with extended-spectrum activity. Diagn Microbiol Infect Dis. 79:373–7. DOI: 10.1016/j.diagmicrobio.2014.03.007. PMID: 24792837.
37. Choi YJ, Kim YA, Junglim K, Jeong SH, Shin JH, Shin KS, et al. 2023; Emergence of NDM-1-producing Pseudomonas aeruginosa sequence type 773 clone: shift of carbapenemase molecular epidemiology and spread of 16S rRNA methylase genes in Korea. Ann Lab Med. 43:196–9. DOI: 10.3343/alm.2023.43.2.196. PMID: 36281514. PMCID: PMC9618910.

38. Hong JS, Song W, Park MJ, Jeong S, Lee N, Jeong SH. 2021; Molecular characterization of the first emerged NDM-1-producing Pseudomonas aeruginosa isolates in South Korea. Microb Drug Resist. 27:1063–70. DOI: 10.1089/mdr.2020.0374. PMID: 33332204.
39. Yoon EJ, Jeong SH. 2021; Mobile carbapenemase genes in Pseudomonas aeruginosa. Front Microbiol. 12:614058. DOI: 10.3389/fmicb.2021.614058. PMID: 33679638. PMCID: PMC7930500. PMID: dd7148a962e44cb78da46857e3ba0ed8.
Fig. 1
Distribution of STs in carbapenem-non-susceptible Pseudomonas aeruginosa isolates collected in three Korean hospitals between 2011 and 2019. (A) STs of P. aeruginosa isolates according to the isolation hospital. (B) STs of P. aeruginosa isolates according to the isolation year.
Abbreviations: ST, sequence type; GNUH, Gyeongsang National University Hospital; KNUH, Kyungpook National University Hospital; JNUH, Jeonbuk National University Hospital.
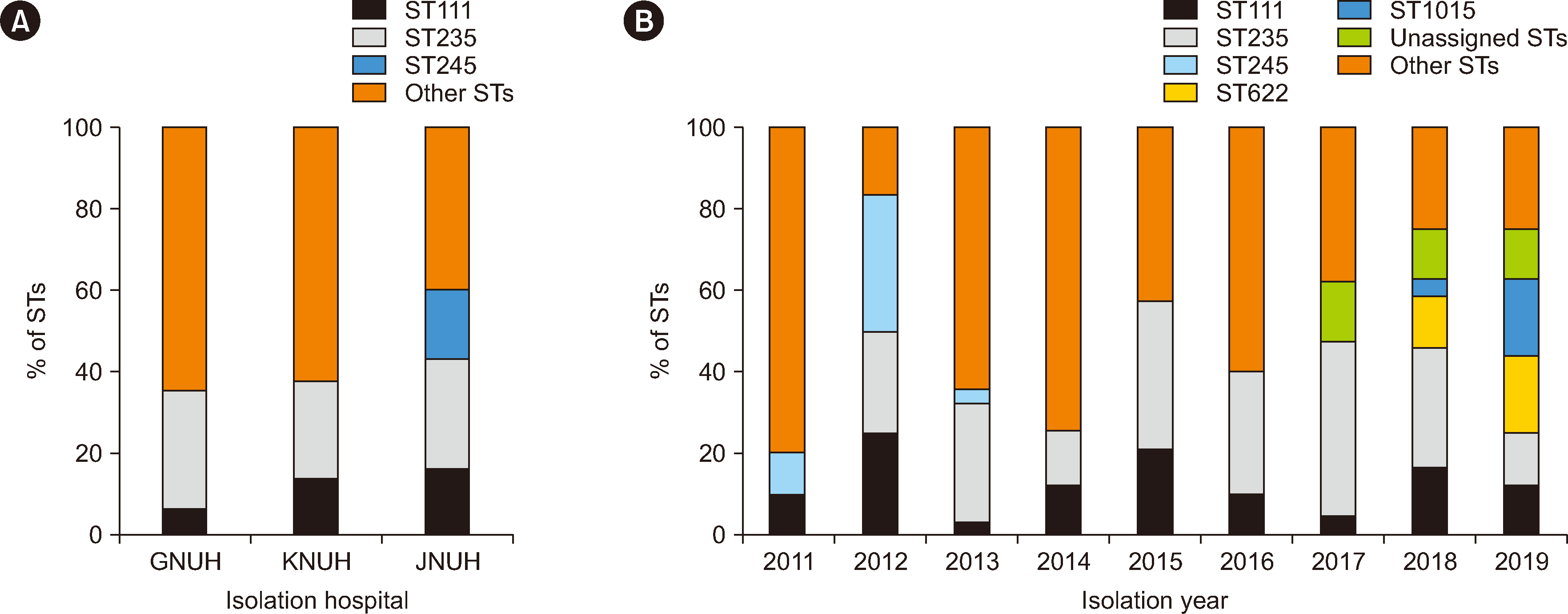
Table 1
Antimicrobial susceptibility of carbapenem-non-susceptible Pseudomonas aeruginosa isolates collected from three Korean hospitals between 2011 and 2019
| Antimicrobial agent | Non-susceptible isolates, N (%) | ||||
|---|---|---|---|---|---|
| ST111 (N=20) | ST235 (N=41) | ST245 (N=10) | Other STs (N=84) | Total (N=155) | |
| Amikacin | 8 (40.0) | 39 (95.1)* | 2 (20.0) | 28 (33.3) | 77 (49.7) |
| Gentamicin | 17 (85.0) | 40 (97.6)* | 8 (80.0) | 49 (58.3) | 114 (73.5) |
| Tobramycin | 16 (80.0) | 40 (97.6)* | 2 (20.0) | 31 (36.9) | 89 (57.4) |
| Piperacillin | 18 (90.0) | 40 (97.6)* | 9 (90.0) | 76 (90.5) | 143 (92.3) |
| Piperacillin/tazobactam | 18 (90.0) | 39 (95.1)* | 9 (90.0) | 75 (89.3) | 141 (91.0) |
| Cefepime | 11 (55.0) | 38 (92.7)* | 7 (70.0) | 65 (77.4) | 121 (78.1) |
| Ceftazidime | 13 (65.0) | 36 (87.8) | 9 (90.0)* | 66 (78.6) | 124 (80.0) |
| Imipenem | 20 (100)* | 41 (100)* | 10 (100)* | 82 (97.6) | 153 (98.7) |
| Meropenem | 18 (90.0) | 41 (100)* | 10 (100)* | 76 (90.5) | 145 (93.5) |
| Aztreonam | 15 (75.0)* | 29 (70.7) | 7 (70.0) | 63 (75.0)* | 114 (73.5) |
| Ciprofloxacin | 18 (90.0) | 40 (97.6)* | 9 (90.0) | 62 (73.8) | 129 (83.2) |
Table 2
Antimicrobial susceptibility of carbapenem-non-susceptible Pseudomonas aeruginosa isolates according to isolation hospital
| Antimicrobial agent | Non-susceptible isolates, N (%) | |||
|---|---|---|---|---|
| KNUH (N=50) | GNUH (N=45) | JNUH (N=60) | Total (N=155) | |
| Amikacin | 27 (54.0)* | 24 (53.3) | 26 (43.3) | 77 (49.7) |
| Gentamicin | 37 (74.0) | 31 (68.9) | 46 (76.7)* | 114 (73.5) |
| Tobramycin | 32 (64.0)* | 23 (51.1) | 34 (56.7) | 89 (57.4) |
| Piperacillin | 45 (90.0) | 42 (93.3)* | 56 (93.3)* | 143 (92.3) |
| Piperacillin/tazobactam | 45 (90.0) | 40 (88.9) | 56 (93.3)* | 141 (91.0) |
| Cefepime | 39 (78.0) | 37 (82.2)* | 45 (75.0) | 121 (78.1) |
| Ceftazidime | 40 (80.0) | 34 (75.6) | 50 (83.3)* | 124 (80.0) |
| Imipenem | 50 (100)* | 43 (95.6) | 60 (100)* | 153 (98.7) |
| Meropenem | 48 (96.0)* | 42 (93.3) | 55 (91.7) | 145 (93.5) |
| Aztreonam | 40 (80.0)* | 30 (66.7) | 44 (73.3) | 114 (73.5) |
| Ciprofloxacin | 38 (76.0) | 38 (84.4) | 53 (88.3)* | 129 (83.2) |
Table 3
Mutations in oprD, carbapenemase genes, expression of ampC, and carbapenem MICs in carbapenem-non-susceptible Pseudomonas aeruginosa isolates
| No. isolate | ST |
Frameshift mutation in oprD (predicted amino acids [N]*) |
Carbapenemase gene | Expression of ampC† | MIC (µg/mL) | |
|---|---|---|---|---|---|---|
| Imipenem | Meropenem | |||||
| KBN10P02278 | 111 | + (0) | - | 2.0 | 32 | 16 |
| KBN12P02599 | 111 | + (241) | - | 0.8 | 16 | 4 |
| KBN06P05035 | 111 | + (441) | - | 1.2 | 8 | 0.5 |
| KBN06P06750 | 111 | + (414) | - | 1.6 | 128 | 16 |
| KBN10P06128 | 235 | + (224) | blaVIM-2 | 1.2 | 256 | 128 |
| KBN10P06311 | 235 | + (93) | blaIMP-6 | 1.3 | 16 | >256 |
| KBN12P04564 | 235 | + (93) | blaIMP-6 | 5.7 | 8 | >256 |
| KBN06P03632 | 235 | + (224) | blaVIM-2 | 4.5 | 128 | 64 |
| KBN06P03793 | 235 | + (93) | blaIMP-6 | 1.4 | 64 | >256 |
| KBN10P04040 | 244 | + (235) | - | 17.2† | 16 | 16 |
| KBN06P05117 | 244 | + (355) | - | 9.5 | 64 | 16 |
| KBN06P02912 | 245 | + (344) | - | 32.9† | 32 | 16 |
| KBN06P03210 | 245 | + (344) | - | 32.8† | 32 | 8 |
| KBN06P02359 | 245 | + (344) | - | 37.5† | 64 | 16 |
| KBN12P06040 | 357 | + (52) | - | 0.2 | 32 | 32 |
| KBN10P06125 | 622 | + (354) | - | 14.7† | 16 | 64 |
| KBN06P06881 | 622 | + (93) | blaIMP-6 | 0.5 | 64 | >256 |
| KBN12P06387 | 1015 | + (235) | - | 1.1 | 16 | 64 |
| KBN10P03382 | 1154 | + (224) | - | 10.2† | 64 | 16 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download