Introduction
Biofilm-associated infections account for 65-80% of all human microbial infections that lead to serious mortality and morbidity [1]. Extracellular polymeric substances (EPS) produced by bacterial or fungal growth protect biofilm-embedded microorganisms against ultraviolet radiation, antibiotics, and disinfection as well as host immune responses [2,3]. A pre-cleaning step using detergents is essential for disinfection and sterilization to remove bioburden that neutralizes the disinfectant [4]. Enzymatic detergents are dispersing biofilm by degradation or hydrolysis of EPS, extracellular DNA, and extracellular proteins within the EPS, however, it cannot eradicate all sort of biofilm or exert bactericidal effect [5,6]. Another type of chemical cleaning facilitator is a non-enzymatic detergent, of which effectiveness is also differed depending on the previous studies [7]. Furthermore, the effectiveness of those varied by the bacterial species of biofilmcausing organism. None of the nine cleaning agents used in the automated cleaning process reached the 4 Log10 reduction in US requirements when applied to endoscopes contaminated with Enterococcus faecium biofilms [8] and that Escherichia coli biofilms are best removed by non-enzymatic detergents [911]. Biofilms caused by Enterococcus faecalis and Pseudomonas aeruginosa are relatively well removed by enzymatic cleaners, but not sufficiently [12]. To date, there is little literature that evaluates the removal ability of medical detergents against biofilms by the species of causative organisms. This study was to compare the efficacy of commercially available enzymatic and non-enzymatic detergents certified for medical use for the biofilm produced by various bacteria and yeasts.
Materials and methods
Study organisms
Type strains and clinical isolates of Escherichia coli, Klebsiella pneumoniae, Citrobacter freundii, Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, Streptococcus agalactiae, Candida albicans, Candida auris, Trichosporon asahii were tested for biofilm production. Laboratory-induced isotypic strains, RN6607 (biofilm production-negative) and RN9120 (biofilm production-positive) of Staphylococcus aureus were included as a control strain [13]. P. aeruginosa ATCC27853, E. coli ATCC25922 and ATCC35218, K. pneumoniae ATCC700603, C. albicans ATCC14053, S. aureus ATCC43300, E. faecalis ATCC29212 were also tested. A total of 33 clinical strains were included in this study; 5 E. coli, 3 K. pneumoniae, 2 C. freundii, 2 P. aeruginosa, 3 A. baumannii, 3 S. aureus, 2 E. faecalis, 2 E. faecium, 3 S. agalactiae, 2 C. albicans, 3 C. auris, and 3 T. asahii. Organisms that produce biofilms were further studied to evaluate the biofilm removal performance of detergents.
Measurement of biofilm production
Biofilm was constructed and quantitated as a previous study [14-17]. Briefly, the organisms suspended in 3% Bacto tryptic soy broth (TSB, Becton Dickinson, Heidelberg, Germany) were cultured overnight at 37°C while shaking at 150 rpm. A 10 μL of culture was diluted to 1:100 in 1 mL of TSB, of which 200 μL were inoculated in polystyrene Falcon 96-well Clear Microplate (#353072, BD Bioscience, Bedford, MA, USA) Tissue-Culture Plates (#353072, Falcon, BD Biosciences, Bedford, MA, USA) and incubated for 24 hr at 37° C. The plates were washed twice with phosphate buffered saline (pH 7.2) and air dried. All tested wells were stained with crystal violet, then were eluted with it with 200 μL of 95% ethanol after washing twice with distilled water (DW). Optical density of tested wells was measured at 620 nm (OD620 ) using EvolisTM system (Bio-Rad, Hercules, CA, USA).
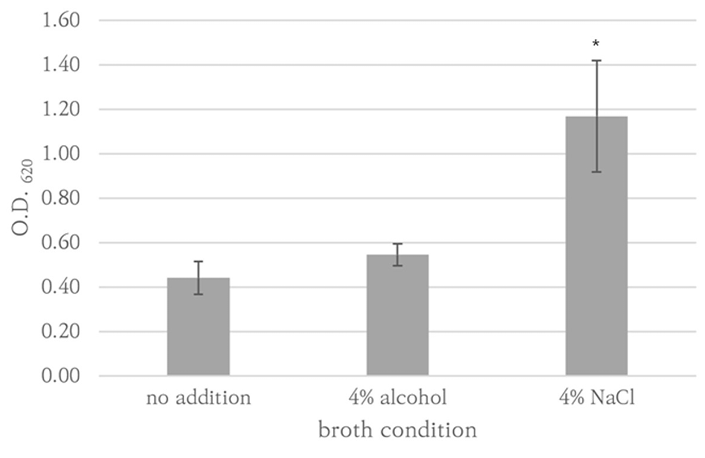
S. aureus RN9120 strain was also tested under conditions in which 4% ethanol or 4% NaCl was added to TSB to enhance biofilm production[13,18]. Each condition was triplicated. Strains showing higher OD620 than S. aureus RN9240 were considered biofilm producers.
Measurement of biofilm removal efficacy
The study detergents included two enzymatic cleaners, EmpowerTM (Metrex, Orange, CA, USA), CidezymeTM (Johnson and Johnson Medical Inc, Arlington, TX, USA), one non-enzymatic cleaner Matrix mintTM (Whiteley Medical, Sydney, Australia). A chlorine bleach YuhanroxTM Regular (Yuhanclorox, Seoul, Korea) was also tested as a control (Table 1). Each biofilm-producing strain was cultured for 72 hr at the same condition of biofilm production to prepare a 96-well microplate coated with biofilm. The biofilmproducing plates prepared by each strain was tested with each detergent under five different conditions: treatment for 8 min once, two times of 8 min, three times of 8 min, and 30 min once at room temperature (RT) and 30 min once at 37℃. After the detergent treatment, OD620 was measured after elution with 100 μ L 95% ethanol in crystal violet-stained wells. The assay value of OD620 was determined by subtracting the value of blank control cultured with TSB medium only from the original value. All OD620 values smaller than the value of blank control were regarded as 0. A single 8-min treatment with DW at RT was taken as the reference for 0% biofilm removal efficacy.
Statistical analysis
For each condition, the mean and standard deviation of three replicates were assigned to OD620 . Using an independent samples t-test (two tailed), biofilm production ability was compared between strains and removal efficacy was compared between detergents. Microsoft Excel (Microsoft Corp., Redmond, WA, USA) was used for statistical analysis.
Results
Biofilm formation of study isolates
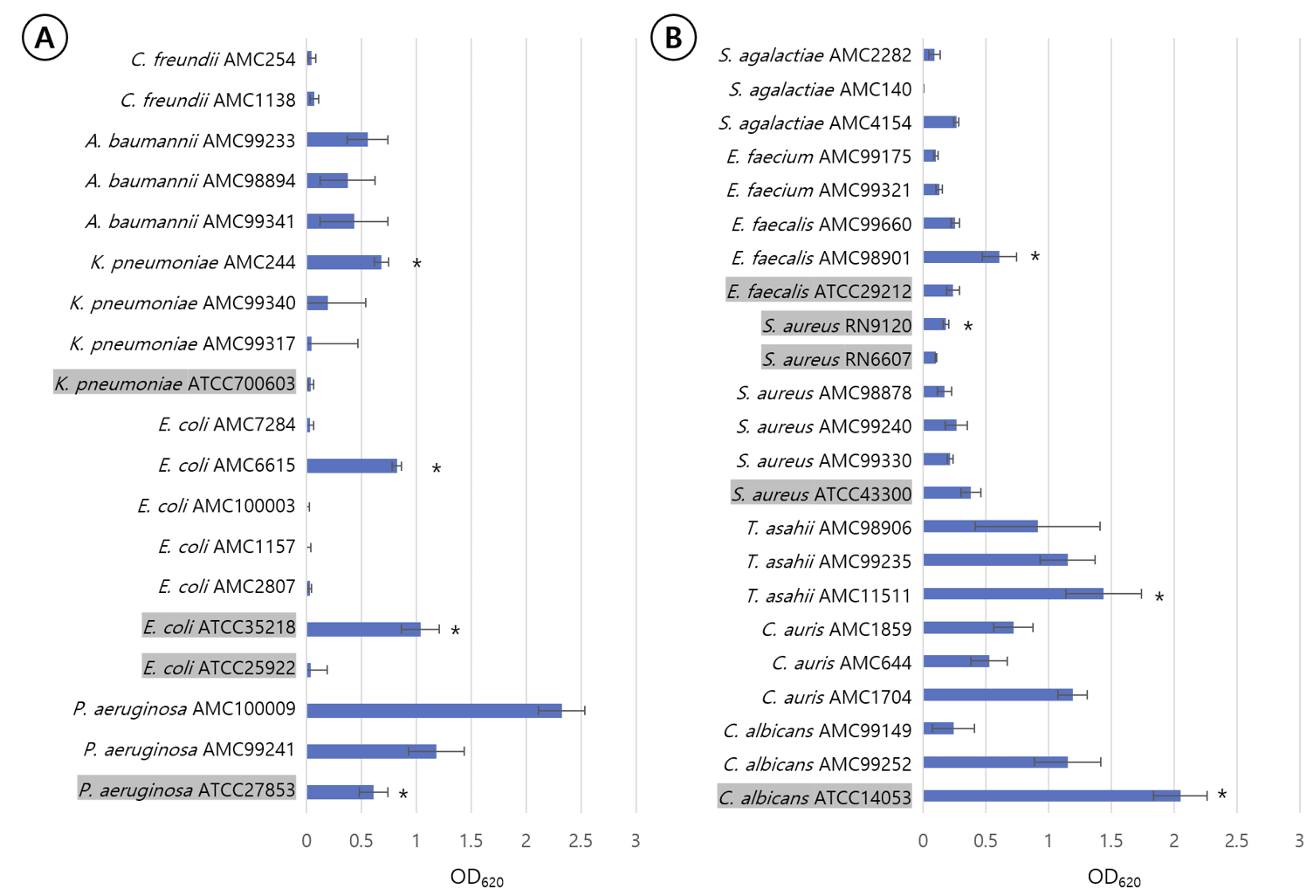
P. aeruginosa ATCC27853, K. pneumoniae AMC244, E. coli ATCC35218, E. coli AMC6615, E. faecalis AMC98901, S. aureus RN9120, C. albicans ATCC14053, C. auris AMC1704, and T. asahii AMC11511 produced significant biofilm production (Fig. 1). S. aureus RN9120 showed an OD620 < 0.5 when cultured with TSB. Because biofilm production of this strain was significantly increased in 4% NaCl-TSB (P = 0.017), but not in 4% ethanol-TSB (P = 0.170) (Fig. 2), it was cultured in 4% NaCl-TSB in biofilm production step when used to test removal efficacy.
Biofilm removal efficacy of three detergents
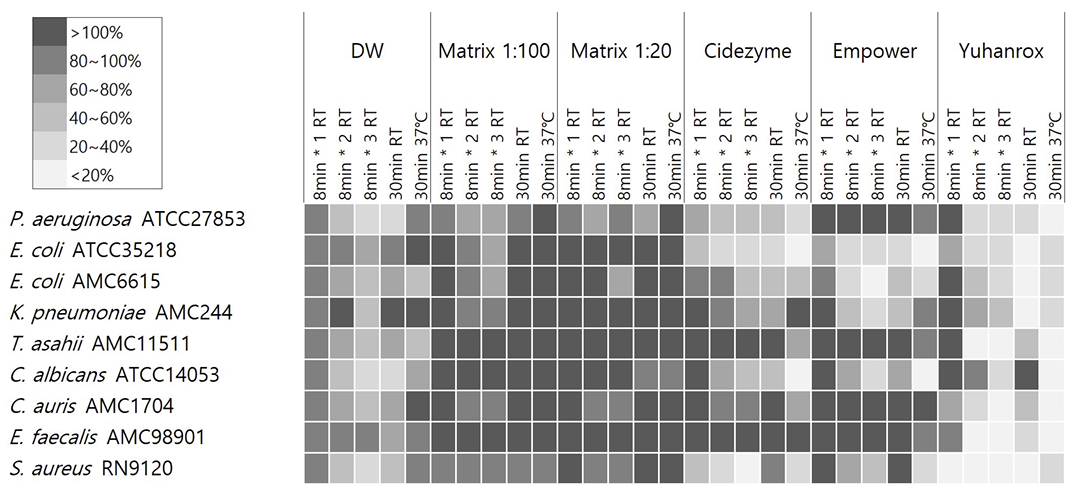
Yuhanrox showed consistent biofilm removal efficacy of > 70% at 37°C incubation for all study isolates. Two enzymatic detergents had biofilm removal rates of greater than 50% for biofilm derived from P. aeruginosa ATCC27853, E. coli ATCC35218, E. coli AMC6615, C. albicans ATCC14053, and S. aureus RN9120 at 37°C for 30 min; 66%, 82%, 52%, 82%, and 71% by Cidezyme, and 13%, 80%, 69%, 92%, and 71% by Empower, respectively. For K. pneumoniae AMC244-derived biofilm, Empower showed the removal rates of 42%, 75%, and 55%, respectively, with treatment for 1, 2, and 30 min at RT, while 5% only when 30 min-treatment at 37°C. None of the detergents were effective enough to remove > 30% of biofilms derived from E. faecalis AMC98901, C. auris AMC1704, and T. asahii AMC11511. Matrix did not show significant removal efficacy against biofilm under any strain or condition (Fig. 3).
Discussion
In this study, P. aeruginosa, C. albicans and T. asahii were the most potent biofilm-producing species, although there was considerable variation among strains in the extent of biofilm production. The results of this study were consistent with previous studies, showing that the ability to produce biofilms varied depending on the strains or conditions [19,20]. P. aeruginosa, C. albicans and T. asahii are opportunistic pathogens as well as well-known causative agents of biofilm-associated infections [21,22]. Biofilm-production ability of strains could be a major virulence factor of the species causing nosocomial infections in healthcare setting. Failure to manually cleaning of medical equipment is often the cause of large-scale nosocomial outbreak [23,24]. Therefore, selecting an effective medical detergent is an important component of control measures of healthcare-associated infections.
The enzymatic detergents showed greater efficacy against biofilms derived from P. aeruginosa, E. coli, C. albicans, and S. aureus, when incubated at 37°C for 30 min. In the previous studies, effect of temperature on biofilm removal efficacy of enzymatic detergents was not established. Although they are often active against biofilm derived from Pseudomonas f luorescens, P. aeruginosa and S. aureus at RT [16,25], they do not eliminate biofilms derived from P. aeruginosa and E. faecalis with 30°C incubation [12]. Because nonenzymatic detergents have activity against EPS, they are expected to be better at removing E. coli biofilms than enzymatic detergents [9-11]. Matrix was not effective for removal of biofilm formed by two E. coli strains under both room temperature and 37°C incubation in this study. Because Matrix is effective against E. coli-derived biofilm when previously tested with clinical isolates [11], the efficacy of Matrix may vary depending on the strain. Enzymatic detergents were more effective than non-enzymatic detergents, especially when treated at 37°C.
All detergents were mostly ineffective against E. faecalis, C. auris, and T. asahii. These results were not surprising because C. auris and T. asahii are well-known yeasts causing biofilm-associated infections and fungal biofilm is usually harder to remove than bacterial biofilm [22,26]. However, the result showing that biofilms derived from E. faecalis were highly resistant to detergents was not consistent with the previous study in which enzymatic detergents were active in removing biofilms derived from E. faecalis ATCC29212 [12]. There may be differences in performance depending on the composition of the detergent used in the study [12]. Alternatively, since more actively biofilm-producing E. faecalis strains were selected among clinical isolates in this study, detergent performance may vary due to the composition of the biofilm matrix produced by the E. faecalis strains. Although there have been efforts to improve effectiveness of biofilm removal [16,27], no commercial detergents have consistently shown reliable activity against biofilms. Therefore, physical cleaning is the only reliable tool in removing biofilms [28,29]. To prevent biofilm-related infections, it is recommended that a combination of chemical cleaning and physical removal are used as a standard method for cleaning and disinfecting medical devices [30].
This study has several limitations. First, the biofilm removal effect of all three detergents was not as good as Yuhanrox and was not consistent by species, strains, and detergent treatment conditions. Therefore, this study did not recommend a detergent that could be used for biofilm removal purposes. Second, although certain strain-treatment combinations were effective in in vitro experiments, it is difficult to predict the efficacy of detergents against actual biofilms. This is because biofilm accumulation occurs in the real world due to the mixing of multiple bacterial and fungal species. [31]. Therefore, it is difficult to predict the efficacy of detergents against actual biofilms through in vitro experiments such as this study.
In conclusion, the biofilm-forming ability of each species or strain of bacteria and yeast varies and biofilmremoval efficacy of detergents is dependent on the strains and detergent treatment condition. Biofilms derived from E. faecalis, C. auris, and T. asahii were resistant to all detergent treatments. No detergent was consistently effective for all microorganisms studied.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download