Abstract
Background: A bloodstream infection is a life-threatening medical emergency, with a mortality rate of up to 30%. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) can be used to identify pathogens directly from positive blood cultures. Two commercial preparation kits, SepsiTyper (Bruker Daltonics, Germany) and SepsiPrep (ASTA Corp., Korea), and two MALDI-TOF MS systems, MALDI Biotyper Sirius (Bruker Daltonics, Germany) and VITEK MS PRIME (bioMérieux, France), are available in Korea. We examined these kits and MALDI-TOF MS systems to analyze their performance. Methods: We assessed the effectiveness of direct identification using 47 blood cultures and 3 bile cultures positive for microbial growth. The VIRTUO system (bioMérieux, France) was used to incubate the samples after they were collected in Bact/ALERT aerobic and anaerobic bottles. The manufacturers’ protocols were followed for both the SepsiTyper and SepsiPrep kits. Results: The SepsiTyper yielded considerably more accurate identifications than did the SepsiPrep, when utilized in MALDI-TOF MS systems (P = 0.0044). However, the SepsiPrep was easier to use and the results more quickly obtained than with the SepsiTyper. The MALDI Biotyper Sirius produced more accurate identifications with the SepsiTyper than did the VITEK MS PRIME (P = 0.0736). The SepsiTyper enabled the accurate identification of five of six polymicrobial cases, utilizing either the MALDI-TOF MS systems. Conclusions: Among the pathogen ID kits tested in this study, the SepsiTyper with MALDI Biotyper Sirius performed the best. In clinical laboratories utilizing VITEK MS PRIME, it is recommended that the either the SepsiTyper or SepsiPrep kit be used for direct identification, while considering certain limitations in terms of performance.
A bloodstream infection (BSI) is a life-threatening medical emergency with a mortality rate of up to 30%, even when treated with empirical antibiotics [1]. Rapid empirical antibiotic treatment is crucial for the treatment of sepsis. Rapid pathogen identification can prevent inappropriate antibiotic treatment, which can improve the emergence of antibiotic resistance [2]. The types of causative bacteria also influence the mortality rate of BSIs. In Korea, the 30-day mortality rates of BSIs caused by Escherichia coli and Klebsiella pneumoniae are 10% and 16.9%, respectively, like in other countries [3]. As a result, accurate and rapid identification of pathogens is crucial not only for proper BSI therapy, but also for predicting prognosis.
Microorganisms can be identified directly from positive blood culture bottles by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) with commercial kits [4–7] or laboratory developed test protocols [8–10]. In Korea, two commercial identification kits are available, the SepsiTyper kit (Bruker Daltonics, Bremen, Germany) and SepsiPrep kit (ASTA Corp., Suwon, Korea), and two MALDI-TOF MS systems, MALDI Biotyper Sirius (Bruker Daltonics, Bremen, Germany) and VITEK MS PRIME (bioMérieux, Marcy l'Etoile, France), are available. Although these MALDI-TOF MS systems have been analyzed using kits from the same manufacturers, their performance with kits from other manufacturers is unknown. There is no straightforward kit for the VITEK MS PRIME (bioMérieux) that includes a lysis buffer and washing solution. In this research, we evaluated the two preparation kits with the two MALDI-TOF MS systems to assess their performance for direct microbial identification.
We examined direct identification procedures for positive blood culture bottles using 50 samples that were positive for microbial growth from September to October 2022 at Dongsan Medical Center. In total, 47 blood samples expected to be monomicrobial and three bile samples expected to be polymicrobial were obtained in Bact/ALERT FA/FN Plus (bioMérieux, Marcy l'Etoile, France) and incubated using the Bact/ ALERT VIRTUO Microbial Detection System (bioMérieux, Marcy l'Etoile, France). When microbial growth was identified by the automatic blood culture system, Gram staining was conducted; monomicrobial cases were defined as those with one, and polymicrobial cases as those with two types of Gram reaction.
The standard SepsiTyper protocol was employed in this study [6]. One milliliter of culture medium from positive blood culture bottles was transferred to an Eppendorf tube and 200 μL of lysis buffer was added. Samples were vortexed for 10–15 seconds, centrifuged at 13,000 rpm for 2 minutes, and the supernatant was discarded. The pellet was centrifuged for 2 minutes at 13,000 rpm, twice, with 1 mL of washing buffe. The supernatant was discarded, and the pellet was washed with 300 μL of distilled water and 900 μL of absolute ethanol by vortexing followed by centrifugation for 2 minutes at 13,000 rpm. The pellet was resuspended in 20 μL of 70% formic acid and 20 μL of acetonitrile, vortexed, then centrifuged for 2 minutes at 13,000 rpm after the supernatant was removed. Next, 1 μL of supernatant was spotted on a MALDI-TOF MS plate, dried for 5 minutes, and 1 μL of the matrix was added. For the MALDI Biotyper Sirius, an identification score ≥ 1.800 indicated species-level identification and scores of 1.600–1.799 indicated genus-level identification. For the VITEK MS PRIME, if the confidence level was ≥ 75% and there was no identification issue, the result was accepted.
One milliliter of culture medium from positive blood culture bottles was transferred to a lysis tube. After vortexing samples for 30 seconds and centrifuging them at 13,000 rpm for 2 minutes, the supernatant was removed. The pellet was washed twice with 1 mL of washing buffer by centrifugation for 2 minutes at 13,000 rpm. After discarding the supernatant and placing the pellet on a MALDI-TOF MS plate, which was then dried for 5 minutes, 1 μL of matrix was added. The scoring criteria of the two MALDI-TOF MS systems were the same as those used with the SepsiTyper kit.
When a microbial growth signal was identified by the BACT/ALERT VIRTUO Microbial Detection System, the sample was subcultured on blood agar and MacConkey agar. Microbial identification using the VITEK MS (bioMérieux) was conducted after 14–48 hours of incubation. The result was approved if the confidence level was 90% or higher and there was identification issue.
The McNcemar chi-squared test was used to evaluate the results of the two systems. The identification of some species within Enterobacter cloacae complex, such as Enterobacter cloacae, Enterobacter hormaechei, Enterobacter bugandensis and Enterobacter kobei, is not accurate when the commercial MALDI-TOF MS system is employed [11]. As a result, if Enterobacter species are identified, regardless of the specific species within the Enterobacter cloacae complex, it was considered an accurate identification at the genus level.
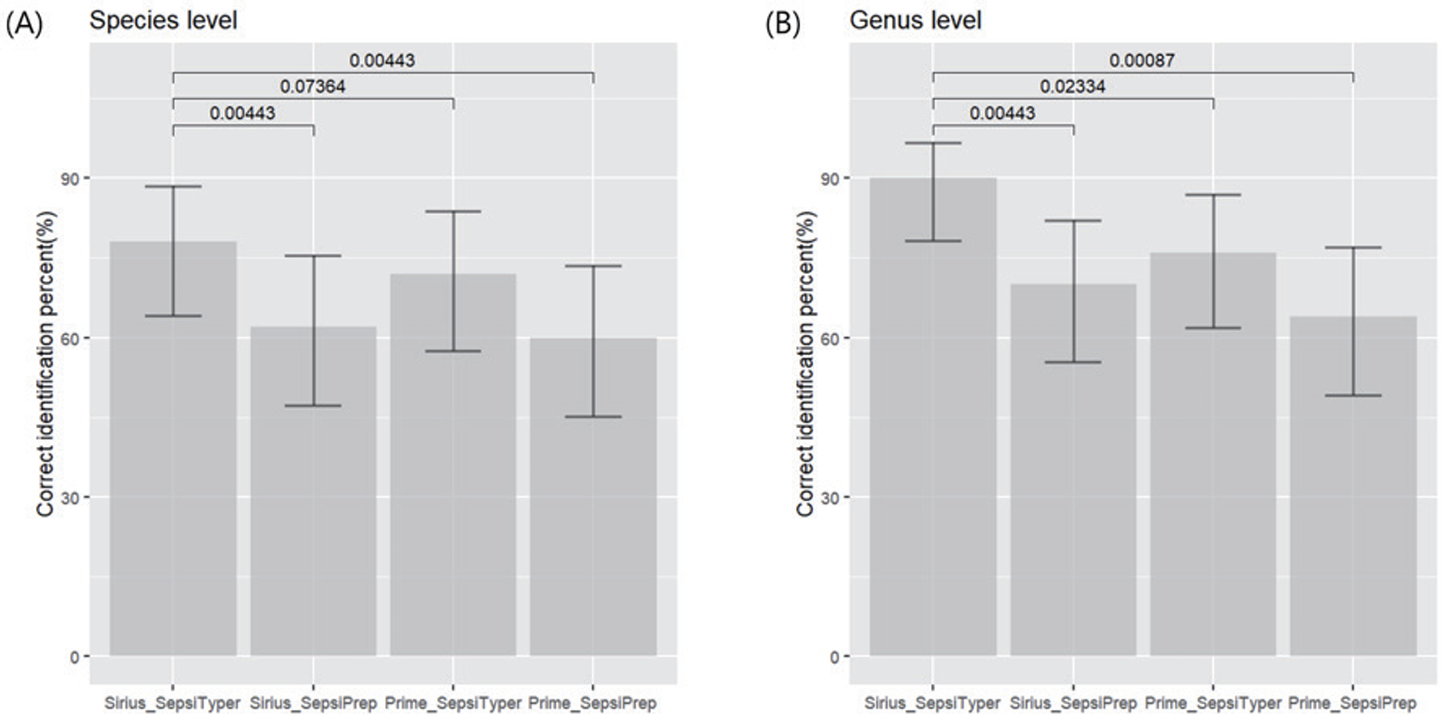
Using the MALDI Biotyper Sirius, the SepsiTyper kit and SepsiPrep kit allowed accurate identification at the species/genus level (including polymicrobial samples in which accurate identification of one or more bacteria was possible at the species/genus level) in 45 (90.0%) and 38 (76.0%) out of 50 total samples, respectively, compared with the conventional identification method (Table 1). The SepsiTyper kit and the SepsiPrep kit both had accurate identification rates at the species level of 78.0% and 58.0%, respectively.
Gram-negative bacteria had greater identification rates at the species/genus levels (100% and 83.3%, SepsiTyper and SepsiPrep kits, respectively) than Gram-positive bacteria (81.8% and 63.6%) and yeasts (100% and 50%), albeit not significantly so (Table 2).
In comparison to the traditional identification method, direct identification with the SepsiTyper kit and SepsiPrep kit allowed the accurate identification of 38 (76.0%) and 32 (64.0%) of 50 isolates at the species/ genus levels, respectively. The accurate identification rate at the species level was 68.0% and 58.0% employing the SepsiTyper kit and SepsiPrep kit, respectively (Table 1). Gram-negative bacteria (88.9% and 77.8% with the Sepsityper and SepsiPrep kits, respectively) had a higher identification rate at the species/genus level than Gram-positive bacteria (68.2% and 59.1%), and yeasts (50% and 50%) (Table 2). The SepsiTyper kit was substantially more accurate than the SepsiPrep kit with both MALDI-TOF MS systems (Fig. 1).
The direct identification outcomes of monomicrobial and polymicrobial cases are listed in Table 3. For monomicrobial and polymicrobial cases, there was no discernible variation in the genus-level correct identification rate.
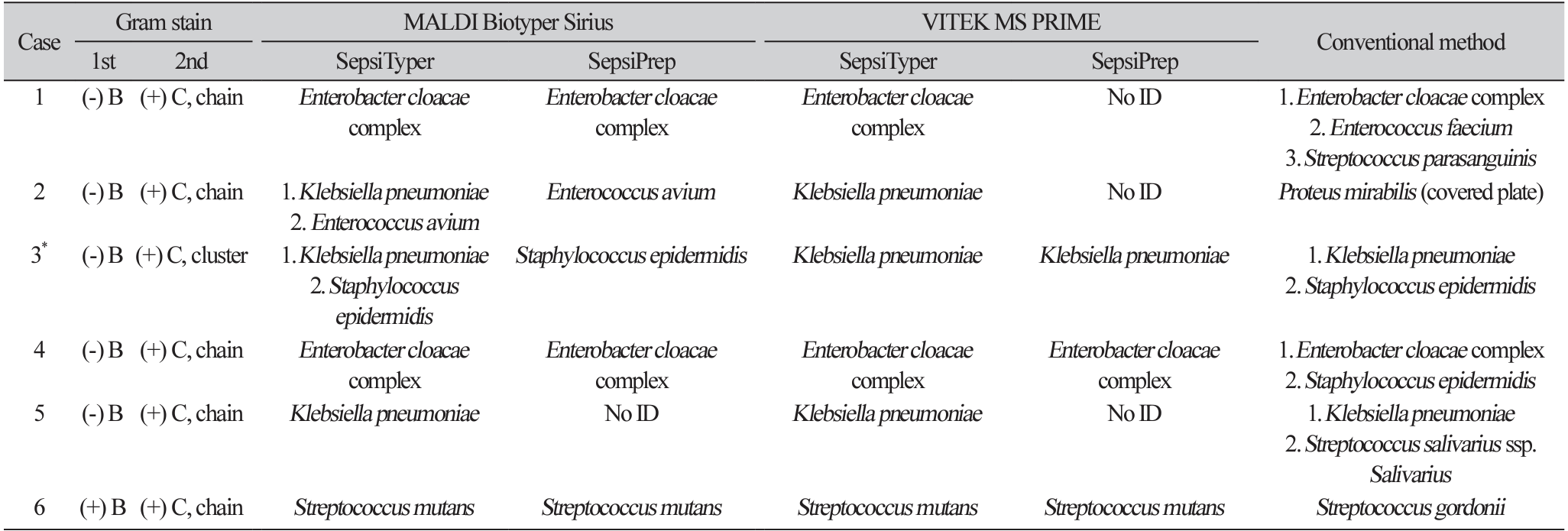
The identification results of the six polymicrobial cases are provided in Table 4. The SepsiTyper kit enabled the accurate identification of five of the six (83.3%) polymicrobial cases at the species/genus level using both MALDI-TOF MS systems (Table 3,4). Polymicrobial cases (66.7% and 50.0%, using MALDI Biotyper Sirius and VITEK MS PRIME, respectively) exhibited a lower identification rate at the species/genus levels than monomicrobial cases (70.5% and 65.9%) with the SepsiPrep kit.
Several risk factors affect the mortality rate of BSIs, which is significant despite empirical antibiotic treatment [1,12–14]. In Japan, age > 65 years, nosocomial infection, qSOFA, SOFA score, Charlson comorbidity index, catheter-related BSI, and urinary tract infection are correlated with the 30-day mortality rate [1]. The varieties of causing bacteria have an impact on BSI death rates as well. Consequently, it is crucial to identify the pathogen responsible for a BSI quickly and accurately. Commercial kits for rapid pathogen identification directly from blood culture bottles by MALDI-TOF MS have been introduced. Their recults vary depending on the commercial kit/protocol utilized, the MALDI-TOF MS equipment, and automated blood culture equipment [10].
The manufacturers of the two commercial kits recommend using each manufacturer’s MALDI-TOF MS system, but there was also a performance testing using different manufacturer’s MALDI-TOF MS system. According to a previous study by Chen et al. [15], the SepsiTyper kit was examined using VITEK MS (old bioMérieux MALDI-TOF MS system) and Bruker Microflex LT (old Bruker Daltonics MALDI-TOF MS system). That study demonstrated that the VITEK MS had a substantially lower identification rate than the Bruker Microflex LT (92.3% and 97.8%, respectively). No study to date has examined the SepsiPrep kit with a MALDI-TOF MS system other than the MicroIDSys Elite (ASTA Corp., Suwon, Korea). In this research, the accurate identification rate of the combination of the SepsiTyper kit and VITEK MS PRIME was 76.0% at the species/genus level, which was lower than the previous report. When utilizing the SepsiPrep kit, the identification rate at the species/genus level was 70.0% in combination with MALDI Biotyper and 64.0% in combination with VITEK MS PRIME. This was the study’s lowest identification rate out of the four kit combinations and MALDI-TOF MS system employed.
In this study, we observed accurate identification rates of 64%-90% using the two preparation kits with the two MALDI-TOF MS systems, similar to previous reports, and the highest identification rate was with the MALDI Biotyper Sirius with the SepsiTyper kit [6,16,17].
With the MALDI Biotyper Sirius, the SepsiTyper kit performed noticeably better than the SepsiPrep kit. The difference was likely a result of various preparation methods. Unlike the SepsiPrep kit, the protocol of the SepsiTyper kit included a formic acid extraction step, improving intracellular protein extraction. The SepsiTyper protocol involved the transfer of the supernatant to the target plate post-extraction, possibly explaining its more consistent results than the SepsiPrep protocol, which involved the spreading of pellets on the target plate. When compared to the SepsiPrep methodology, which only took 10 minutes to complete a test, the SepsiTyper protocol took over 30 minutes [5]. All components of the SepsiPrep kit were provided lyophilized, simplifying the process.
The combination of the SepsiTyper kit with the MALDI Biotyper Sirius revealed significantly superior performance to the VITEK MS PRIME. The SepsiTyper kit is made by the same company that makes the MALDI Biotyper Sirius, which is one of the main causes of these variations. Additionally, VITEK MS PRIME does not have a separate database and algorithms for the direct identification of pathogens, such as the MBT-SepsiTyper module of the MALDI Biotyper Sirius. To minimize these performance variations, direct identification reagents created for VITEK MS PRIME and soft software updates are required.
The reported identification rates of Gram-negative bacteria are 31.4%-100%, and those of Gram-positive bacteria are 54.5%-92.9% [4,10,15–18]. In this study, the identification rates were 59.1%-81.8% for Grampositive bacteria and 77.8%-100% for Gram-negative bacteria. This might be because Gram-positive bacteria have a thick peptidoglycan layer, which makes it challenging to detect intracellular proteins [4]. The low identification rate of yeast may be described similarly.
At least one organism was identified in 50% of the polymicrobial cases using both MALDI-TOF MS systems with the SepsiTyper kit. In a previous study, one organism was identified from all polymicrobial samples, which could be enhanced by software updates [19]. Of the six polymicrobial samples in this study, both strains were identified in one sample using the MALDI Biotyper Sirius with the SepsiTyper kit (Table 3, case No. 3). In case 2, Gram-positive cocci in chains and Gram-negative bacilli were observed, and Proteus mirabilis covered the agar surface by swarming. Therefore, bacteria with Gram-positive cocci chains and Gram-negative bacilli could not be identified by the conventional method.
This study had several drawbacks. First, we had a small number of specimens, especially of yeasts and anaerobic bacteria. The identification rate of yeast using the MBT-SepsiTyper module was lower than that of Gram-positive and -negative bacteria (65.4% and 78.9%, respectively), while that of anaerobic bacteria was lower than that of aerobic bacteria (76.2% and 61.9%, respectively) [6]. Second, we could not utilize the MALDI-TOF MS system from ASTA Corp. However, the identification rate using the SepsiPrep kit and MicroIDSys Elite was 96.5% and 98.5% for Gram-positive and -negative isolates, respectively [5]. Third, we could not use the VITEK MS Blood culture kit (bioMérieux, Marcy l'Etoile, France), which, in conjunction with the VITEK MS, has an identification rate of 73% [9]. Because the protocol involves lysisf iltration, additional facilities are needed [9]. Fourth and finally, we could not conduct a direct antimicrobial susceptibility test (AST) study with automated AST instruments. Some commercial, automated AST instruments yielded AST results associated with those of conventional AST using pellets derived from subcultured colonies [5].
This study is the first to examine two preparation kits with two MALDI-TOF MS systems in clinical laboratories in Korea. The SepsiTyper kit with MALDI Biotyper Sirius revealed the best performance. Currently, since bioMérieux's preparation kit is not readily available in Korea, clinical laboratories using VITEK MS PRIME may consider using the SepsiTyper kit or SepsiPrep kit. However, given the clear performance differences found in this study, it is important to have a enough understanding of their limitations for direct identification.
This multicenter study provides evidence of asymptomatic colonization by Blastocystis, particularly ST3, in the gastrointestinal tracts of individuals in Korea. Our findings also reveal regional differences in the distribution of Blastocystis and its subtypes. These results underscore the need for further research to elucidate the potential impact of Blastocystis on human health.
Ethics statement
This study was approved by the Institutional Review Board of Dongsan Medical Center (IRB No. 202211-080).
REFERENCES
1. Hattori H, Maeda M, Nagatomo Y, Takuma T, Niki Y, Naito Y, et al. Epidemiology and risk factors for mortality in bloodstream infections: a single-center retrospective study in Japan. Am J Infect Control 2018;46:e75–9.
.
2. Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intens Care Med 2021;47:1181–247.
.
3. Yoon EJ, Kim D, Jeong SH. Bloodstream infections and carbapenem-resistant Enterobacteriaceae in South Korea. Lancet Infect Dis 2019;19:931–2.
.
4. Fre´de´ric S, Antoine M, Bodson A, Lissoir B. Bacterial rapid identification with matrix assisted laser desorption/ionization time-of-flight mass spectrometry: development of an ‘inhouse method’ and comparison with Bruker Sepsityper® kit. Acta Clin Belg 2015;70:325–30.
.
5. Yoo IY, Han J, Ha SI, Cha YJ, Pil SD, Park Y. Clinical performance of ASTA SepsiPrep kit in direct bacterial identification and antimicrobial susceptibility test using MicroIDSys Elite and VITEK-2 system. J Clin Lab Anal 2021;35:e23744.
.
6. Ponderand L, Pavese P, Maubon D, Giraudon E, Girard T, Landelle C, et al. Evaluation of Rapid Sepsityper® protocol and specific MBT-Sepsityper module (Bruker Daltonics) for the rapid diagnosis of bacteremia and fungemia by MALDI-TOF-MS. Ann Clin Microbiol Antimicrob 2020;19:60.
.
7. Lin HH, Tseng KH, Tien N, Lin YT, Yu J, Hsueh PR, et al. Evaluation of the Rapid Sepsityper protocol and specific MBT-Sepsityper module for the identification of bacteremia and fungemia using Bruker Biotyper MALDI-TOF MS. J Microbiol Immunol Infect 2022;55:1330-3.
.
8. Pranada AB, Cordovana M, Meyer M, Hubert H, Abdalla M, Ambretti S, et al. Identification of micro-organism from positive blood cultures: comparison of three different short culturing methods to the Rapid Sepsityper workflow. J Med Microbiol 2022;71:001571.
.
9. Fothergill A, Kasinathan V, Hyman J, Walsh J, Drake T, Wang YFW. Rapid identification of bacteria and yeasts from positive-blood-culture bottles by using a lysis-filtration method and matrix-assisted laser desorption ionization-time of flight mass spectrum analysis with the SARAMIS database. J Clin Microbiol 2013;51:805–9.
.
10. Morgenthaler NG and Kostrzewa M. Rapid identification of pathogens in positive blood culture of patients with sepsis: review and meta-analysis of the performance of the Sepsityper kit. Int J Microbiol 2015;2015:1-10.
.
11. Godmer A, Benzerara Y, Normand AC, Veziris N, Gallah S, Eckert C, et al. Revisiting species identification within the Enterobacter cloacae complex by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Microbiol Spectr 2021;9:e00661-21.
.
12. Brady M, Oza A, Cunney R, Burns K. Attributable mortality of hospital-acquired bloodstream infections in Ireland. J Hosp Infect 2017;96:35–41.
.
13. Dat VQ, Vu HN, Nguyen The H, Nguyen HT, Hoang LB, Vu Tien Viet D, et al. Bacterial bloodstream infections in a tertiary infectious diseases hospital in Northern Vietnam: aetiology, drug resistance, and treatment outcome. BMC Infect Dis 2017;17:493.
.
14. Goto M and Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 2013;19:501–9.
.
15. Chen JHK, Ho PL, Kwan GSW, She KKK, Siu GKH, Cheng VCC, et al. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J Clin Microbiol 2013;51:1733–9.
.
16. Schieffer KM, Tan KE, Stamper PD, Somogyi A, Andrea SB, Wakefield T, et al. Multicenter evaluation of the SepsityperTM extraction kit and MALDI-TOF MS for direct identification of positive blood culture isolates using the BD BACTECTM FX and VersaTREK® diagnostic blood culture systems. J Appl Microbiol 2014;116:934–41.
.
17. Jamal W, Saleem R, Rotimi VO. Rapid identification of pathogens directly from blood culture bottles by Bruker matrix-assisted laser desorption laser ionization-time of flight mass spectrometry versus routine methods. Diagn Microbiol Infect Dis 2013;76:404–8.
.
18. Prod’hom G, Bizzini A, Durussel C, Bille J, Greub G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J Clin Microbiol 2010;48:1481–3.
.
19. Tadros M and Petrich A. Evaluation of MALDI-TOF mass spectrometry and Sepsityper Kit™ for the direct identification of organisms from sterile body fluids in a Canadian pediatric hospital. Can J Infect Dis Med Microbiol 2013;24:191–4.
.
Fig. 1.
Accurate identification rates for each combination of reagent kit and MALDI-TOF MS system at the species (A) and genus (B) levels. MALDI-TOF MS, matrix-assisted laser desorption/ionization timeof-flight mass spectrometry.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download