Abstract
Background: Blastocystis is a genus of intestinal, anaerobic protozoan parasites that can be isolated from humans, animals, and the environment. We aimed to determine the distribution of Blastocystis and subtypes (STs) using stool samples obtained from healthy volunteers at collection centers in Korea. Methods: A total of 478 stool samples from volunteers were collected at five collection centers throughout Korea. The presence of Blastocystis was determined using PCR based on the small subunit (SSU) rRNA gene, and Blastocystis STs were confirmed through sequencing of the SSU rRNA gene. Results: Molecular analysis revealed the presence of Blastocystis in 27 (5.6%) of the enrolled participants. Two STs were identified: ST3 (66.7%) and ST1 (33.3%). The positive rates of Blastocystis varied by geographical region, ranging from 1.2%–12.0%. ST3 was the predominant subtype in all centers except one, where only ST1 was isolated. Phylogenic analysis showed clustering based on ST, but no significant differences were found among the regions. There was no association between Blastocystis colonization and either age or sex of the participants. Conclusions: The results of this multicenter study demonstrated colonization by Blastocystis, mainly ST3, in the gastrointestinal tracts of asymptomatic individuals in Korea.
Blastocystis–a genus of intestinal, anaerobic protozoan parasites–has recently been recognized as the most prevalent eukaryotic microbe colonizing the human gut [1-3]. It is thought to be transmitted via the fecal–oral route in the cyst form [4]. Numerous epidemiological studies have highlighted the global distribution of Blastocystis sp., with high prevalence of this protozoan infection in developing countries. Recent advances in molecular approaches have shed light on the distribution, pathogenicity, and genetic diversity of Blastocystis [5]. Of the reported 23 distinct subtypes (STs) [6], Blastocystis ST1–ST9 and ST12 have been identified in humans, with ST1–ST4 being the most common [7]. Collectively, ST1–ST4 comprise more than 90% of all identified STs, with ST3 being the most common in humans [7]. Despite being one of the most widely studied protozoans, knowledge about the biology and pathophysiology of Blastocystis is still limited. To date, there exist only two published epidemiological reports of human Blastocystis infection or coloniazation among Koreans [8,9]. In this study, we conducted a multicenter investigation of the prevalence of Blastocystis and their STs among asymptomatic individuals in Korea.
A total of 478 stool samples from volunteers were collected at four university hospitals (Chonnam National University Hwasun Hospital, Gyeongsang National University Hospital, Hanyang University Guri Hospital, and Gangneung Asan Medical Center) and Green Cross Laboratories from July 2021 through March 2022. Fecal samples were collected in accordance with the guidelines of the Institutional Review Board of Chonnam National University Hwasun Hospital (IRB No. CNUHH-2022-112). DNA was extracted using the Cica Geneus DNA Prep Kit (Kanto Chemical, Tokyo, Japan) according to the manufacturer’s instructions. Blastocystis was detected based on the small subunit ribosomal RNA gene using Blast-505-532 (5′–GGA GGT AGT GAC AAT AAA TC–3′) and Blast-998-1017 (5′–TGC TTT CGC ACT TGT TCA TC–3′) primers [8]. Each tube contained 8.5 μL of solution including the pair of polymerase chain reaction (PCR) primers (1 μL each of Blast-505-532 and Blast-998-1017 at 25 pmol), 36.5 μL of distilled water, and 5 μ L of template DNA. All PCR amplifications were performed using the TaKaRa PCR Thermal Cycler Dice Gradient (TaKaRa, Tokyo, Japan). The conditions used were as follows: initial denaturation at 94°C for 3 min, followed by 30 cycles of 59°C for 30 s and 72°C for 60 s, and a final extension at 72°C for 5 min. The PCR products were analyzed using 1.5% agarose gel electrophoresis with ethidium bromide staining and then sent to Macrogen (Seoul, Korea) for direct DNA sequencing. Phylogenetic analysis was performed using the sequences of Blastocystis SSU rRNA gene, and a phylogenetic tree was constructed using Geneious Prime (Biomatters Ltd, Auckland, New Zealand). Phylogenetic inference was conducted using the unweighted pair group method with arithmetic mean clustering with 1,000 bootstrap replications.
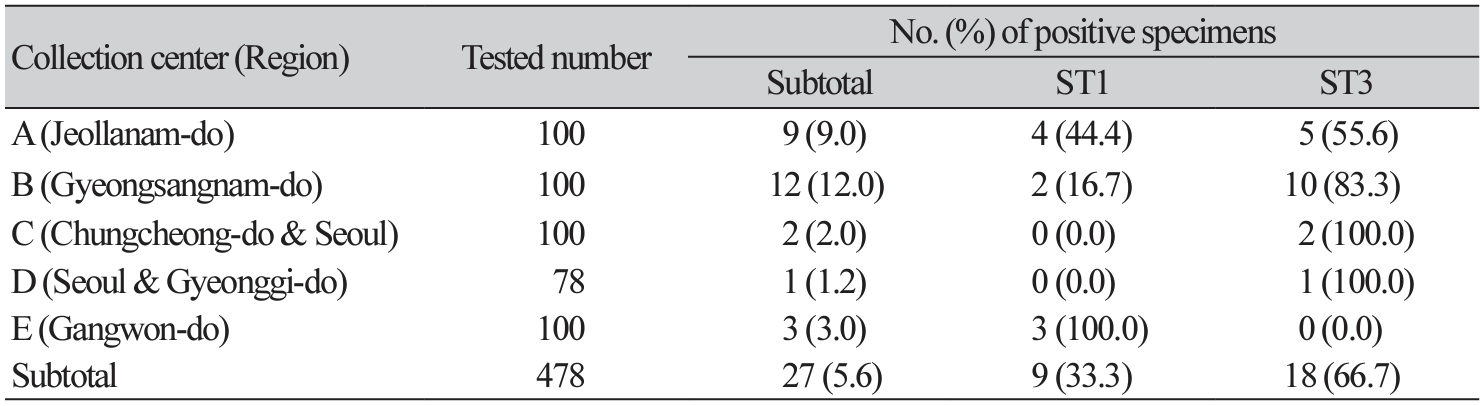
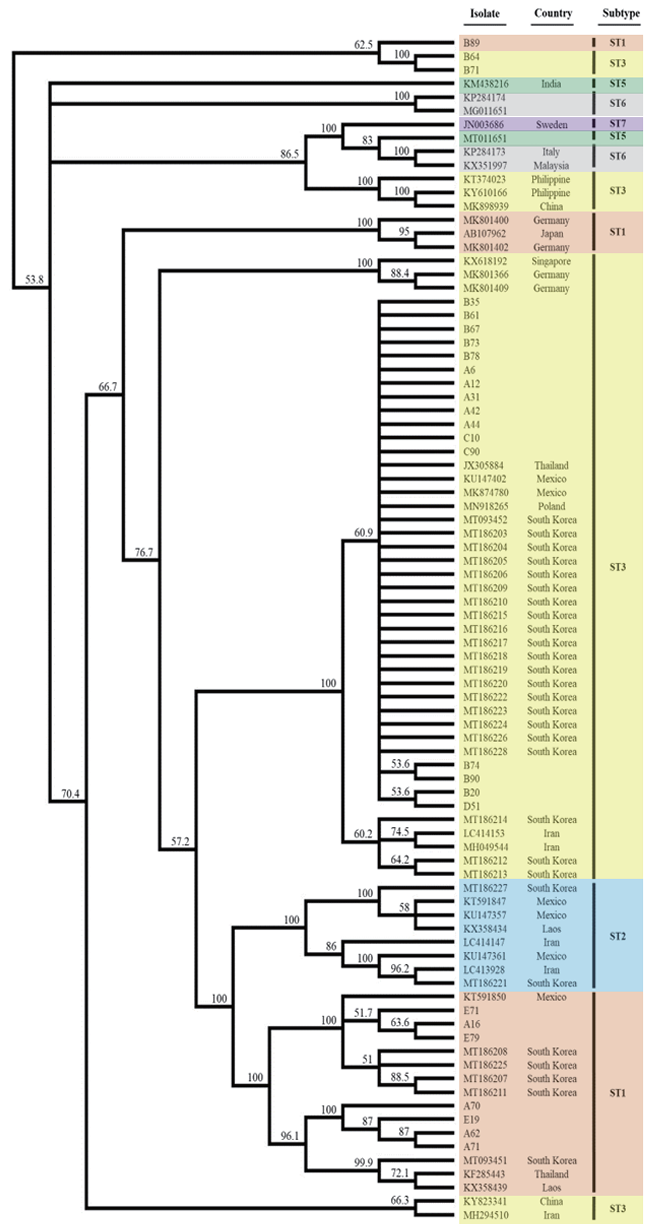
Overall, 27 (5.6%) of the enrolled 478 participants was positive for Blastocystis (Table 1). The wide range of positivity rates were noticed from 1.2%–12.0% depending on geographical region of the participating institutions. However, there were no significant differences in age or gender among the participating institutions (data not shown). ST3 (66.7%) was the most common Blastocystis subtype, and the other subtype identified was ST1 (33.3%). ST3 was the predominant subtype at four centers (centers A-D), while the only ST1 was found at the center located in Gangwon-do. Our phylogenic tree analysis showed ST clustering and there were no significant differences among the regions in this regard (Fig. 1). All but two sequences of ST3 were co-clustered with sequences from other countries, such as Thailand (JX305884), Mexico (KU147402, MK874780), and Poland (MN918265), in addition to previously reported Korean strains (MT093452, MT186203-MT186206, MT186209, MT186210, MT186215-MT186228). Of note, eight ST1 sequences were divided into three groups consisting of four isolates (A62, A70, A71, E19), three isolates (A16, E71, E79), and one isolate (B89). One isolate collected from collection center B was distant from the other isolates collected from collection centers A and E.
The pathogenicity of Blastocystis is still unclear and debate over whether it is a pathogen or mutualist should consider environmental, dietary, and geographic factors [10]. In a previous study, the positive rate of Blastocystis was significantly lower in the diarrheal group (3.1%) than in the non-diarrheal group (18.0%) [8], suggesting that colonization of Blastocystis may be frequent. Here, we confirmed that the Blastocystis is frequently colonized among non-symptomatic population in Korea. This is consistent with a previous metaanalysis that demonstrated a pooled human blastocystosis prevalence of 9.1% (range, 0.01%–35.2%) [11]. In several Asian studies, prevalences of Blastocystis infection have been determined as follows: 11.6% in China, 49.1% in the Philippines, 34.3% in Indonesia, and 22.3% in Thailand [11]. In this study, we also noticed significantly varying Blastocystis positivity rates according to geographical region, ranging from 1.2%–12.0%. Of note, two facilities located in southern Korea had relatively high prevalences (11.0%–12.0%), while the other centers had low prevalences (1.2%–3.0%). Epidemiological studies have often found that Blastocystis prevalence usually vary by geographical region and that there is a tendency for individuals carrying Blastocystis to be inhabitants of areas with inadequate sanitation [3,12,13]. Chang et al.[9] previously found a low frequency of cooked or boiled vegetable consumption to be a potential risk factor for Blastocystis spp. colonization. Javanmard et al. [11] demonstrated a close correlation between blastocystosis prevalence and the climate and hygiene conditions of investigated populations. These observations may partly explain the two centers with high blastocystosis prevalence in our study: these are located in rural areas and are associated with a high probability of frequent raw vegetable consumption.
In our study, only two STs�ST3 (66.7%) and ST1 (33.3%)�were identified. This finding is similar to that reported in China, as well as North and South America, where high prevalence of ST3 have been observed, followed by ST1 and ST2 [14]. Regarding Korean epidemiology, Kim et al.[8] first reported ST3 to be the dominant type in southern Korea, although it was more frequently observed in people without diarrhea than people with diarrhea. Subsequently, a cross-sectional study targeting older adults undergoing health checkups at the Seoul Western Branch of the Korea Association of Health Promotion was performed and also confirmed ST3 and ST1 as the two major Blastocystis STs [9]. However, those two studies were limited to single centers, respectively. Our multicenter study demonstrated colonization by Blastocystis, mainly ST3, in the gastrointestinal tracts of asymptomatic people in Korea. Notably, we observed ST variations according to geographic region. ST3 was the major ST at all but one center, and ST1 was only isolated from specimens collected at the center located in Gangwon-do. Kim et al.[8] previously suggested ST1 to be correlated with diarrheal symptoms, unlike ST3. It has been reported that Blastocystis sp. can have both beneficial and harmful effects depending on the richness and species composition of the host gut microbiota, as well as the associated abundance and composition of Blastocystis STs [15]. It is important to identify certain STs among Blastocystis sp. because clinical symptoms, such as diarrhea and irritable bowel syndrome, may be associated with certain STs [16]. On the other hand, a very recent data announced that there was no significant effect of short-term exposure to Blastocystis ST3 on gut inflammation following colitis induction, while longterm exposure appeared to promote a faster recovery from colitis [17]. Continuous studies should be further conducted to expand on clinical implication of Blastocystis.
This multicenter study provides evidence of asymptomatic colonization by Blastocystis, particularly ST3, in the gastrointestinal tracts of individuals in Korea. Our findings also reveal regional differences in the distribution of Blastocystis and its subtypes. These results underscore the need for further research to elucidate the potential impact of Blastocystis on human health.
Ethics statement
This study was approved by the Institutional Review Board of Chonnam National University Hwasun Hospital (IRB No. CNUHH-2022-112).
Conflicts of interest
Jung-Hyun Byun is currently an Assistant Editor of the Annals of Clinical Microbiology. However, she was not involved in the review process of this article.
Acknowledgements
We thank all members of the Tropical Medicine Research Group of the Korean Society of Clinical Microbiology.
Funding
This work was partially supported by the Research Fund (2021) of the Korean Society of Clinical Microbiology, as well as the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2022R1C1C1002741).
REFERENCES
1. Wawrzyniak I, Poirier P, Viscogliosi E, Dionigia M, Texier C, Delbac F, et al. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther Adv Infect Dis 2013;1:167–78.
.
2. Parija SC and Jeremiah S. Blastocystis: taxonomy, biology and virulence. Trop Parasitol 2013;3:17–25.
.
3. Stensvold CR and Clark CG. Current status of Blastocystis: a personal view. Parasitol Int 2016;65:763–71.
.
4. Lepczyńska M, Białkowska J, Dzika E, Piskorz-Ogórek K, Korycińska J. Blastocystis: how do specific diets and human gut microbiota affect its development and pathogenicity? Eur J Clin Microbiol Infect Dis 2017;36:1531–40.
.
5. Rahimi HM, Pourhosseingholi MA, Yadegar A, Mirjalali H, Zali MR. High-resolution melt curve analysis: a real-time based multipurpose approach for diagnosis and epidemiological investigations of parasitic infections. Comp Immunol Microbiol Infect Dis 2019;67:101364.
.
6. Stensvold CR and Clark CG. Pre-empting Pandora's box: Blastocystis subtypes revisited. Trends Parasitol 2020;36:229–32.
.
7. Popruk S, Adao DEV, Rivera WL. Epidemiology and subtype distribution of Blastocystis in humans: a review. Infect Genet Evol 2021;95:105085.
.
8. Kim MJ, Won EJ, Kim SH, Shin JH, Chai JY. Molecular detection and subtyping of human Blastocystis and the clinical implications: comparisons between diarrheal and non-diarrheal groups in Korean populations. Korean J Parasitol 2020;58:321–6.
.
9. Chang T, Jung BK, Shin H, Hong S, Ryoo S, Lee J, et al. Genotypes of Blastocystis sp. among elderly health checkup people in South Korea with a questionnaire on risk factors. Parasitol Res 2021;120:3297–306.
.
10. Kim MJ, Lee YJ, Kim TJ, Won EJ. Gut microbiome profiles in colonizations with the enteric protozoa Blastocystis in Korean populations. Microorganisms 2021;10:34.
.
11. Javanmard E, Niyyati M, Ghasemi E, Mirjalali H, Aghdaei HA, Zali MR. Impacts of human development index and climate conditions on prevalence of Blastocystis: a systematic review and meta-analysis. Acta Trop 2018;185:193–203.
.
12. Belleza ML, Cadacio JL, Borja MP, Solon JA, Padilla MA, Tongol-Rivera PN, et al. Epidemiologic study of Blastocystis infection in an urban community in the Philippines. J Environ Public Health 2015;2015:894297.
.
13. Poulsen CS, Efunshile AM, Nelson JA, Stensvold CR. Epidemiological aspects of Blastocystis colonization in children in Ilero, Nigeria. Am J Trop Med Hyg 2016;95:175–9.
.
14. Jiménez PA, Jaimes JE, Ramírez JD. A summary of Blastocystis subtypes in North and South America. Parasit Vectors 2019;12:376.
.
15. Deng L, Wojciech L, Gascoigne NRJ, Peng G, Tan KSW. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog 2021;17:e1009253.
.
16. Moosavi A, Haghighi A, Mojarad EN, Zayeri F, Alebouyeh M, Khazan H, et al. Genetic variability of Blastocystis sp. isolated from symptomatic and asymptomatic individuals in Iran. Parasitol Res 2012;111:2311–15.
.
17. Billy V, Lhotská Z, Jirků M, Kadlecová O, Frgelecová L, Parfrey LW, et al. Blastocystis colonization alters the gut microbiome and, in some cases, promotes faster recovery from induced colitis. Front Microbiol 2021;12:641483.
.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download