Abstract
Background: We evaluated the diagnostic performance of the Exdia COVID-19 antigen test (Exdia Ag; Precision Biosensor Inc., Korea) as a point-of-care (POC) test performed in the emergency department (ED) for the rapid detection of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in comparison with the performance of the Real-Q 2019-nCoV Detection KIT (Real-Q; BioSewoom, Korea). Methods: Exdia Ag and Real-Q assays were performed simultaneously for all patients who were admitted to the ED of Seoul St. Mary's Hospital with or without COVID-19 symptoms between December 2021 and March 2022. Results: Among the 2,523 samples analyzed by Real-Q assay, 149 samples (5.9%) showed positive results, and 2,374 samples showed negative results. The overall sensitivity and specificity of the Exdia Ag assay were 77.2% (95% confidence interval [CI], 69.6 – 83.7) and 99.8% (95% CI, 99.6 – 99.9), respectively. The positive and negative predictive values were 96.6% (95% CI, 91.5 – 98.7) and 98.6% (95% CI, 98.1 – 98.9), respectively. The cycle threshold value for 115 concordant Exdia Ag-positive/Real-Q-positive samples was significantly lower than that for 34 discordant Exdia Ag-negative/Real-Q-positive samples (P< 0.0001). Conclusion: The Exdia Ag assay showed good diagnostic performance when the disease prevalence was high and may be useful for POC testing when the rapid detection of SARSCoV-2 is required for the isolation of patients in the ED.
As the prevalence of coronavirus disease 2019 (COVID-19) gets higher, sophisticated test strategies are required in different settings. Although highly sensitive and specific, real-time reverse-transcription polymerase chain reaction (rRT-PCR) is the reference method for diagnosing COVID-19 [1,2], as it takes several hours, more rapid diagnostic tests are needed especially in the emergency department (ED). Therefore, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) antigen tests have emerged as pointof-care (POC) testing which provide results within 15 – 30 minutes, leading to timely detection of infectious patients in the ED [3]. During the COVID-19 pandemic in Korea, early detection, and proper allocation of COVID-19 patients in ED can be achieved by using POC SARS-CoV-2 antigen test. There were several reports that the POC SARS-CoV-2 antigen test was implemented in ED to help patients in timely isolation [4-6]. Here, we evaluated the Exdia COVID-19 antigen test (Exdia Ag; Precision Biosensor Inc., Daejeon, Korea) as a POC testing performed in ED compared to the Real-Q 2019-nCoV Detection KIT (Real-Q; BioSewoom, Seoul, Korea) between December 2021 and March 2022.
For all patients who admitted to the ED of Seoul St. Mary's Hospital with or without COVID-19 symptoms, we conducted both Exdia Ag and Real-Q assay simultaneously. For each patient, one nasopharyngeal swab for Exdia Ag assay and both oropharyngeal and nasopharyngeal swabs for Real-Q assay were collected by trained medical personnel (doctors, nurses, and technicians). For Exdia Ag assay, nasopharyngeal swab was mixed with the diluent by immersing into the diluent tube. Then, two drops were delivered to the sample-loading place in the cassette. Then, the cassette was inserted into Exdia TRF Plus analyzer (Precision Biosensor Inc.) and result was interpreted automatically by the signal reader as positive, negative, and invalid result (absence of a control line) within 20 minutes. For rRT-PCR, oropharyngeal and nasopharyngeal swabs were collected in the virus transport medium (VTM; Noble Bio, Hwaseong, Korea), and RNA extraction was carried out using the NC-15 PLUS (Hanwool TPC, Bucheon, Korea). The extracted RNA was used for Real-Q assay using the real-time PCR instrument CFX96 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Real-Q assay targets regions of envelope (E gene) and RNA dependent RNA polymerase (RdRp gene) of SARS-CoV-2. The cycle threshold (Ct) value < 38 for all two target genes was defined as a positive result. If one target gene is positive, but the other is negative, the test is considered indeterminate.
Sensitivity, specificity, positive predictive values (PPVs) and negative predictive values (NPVs) with 95% confidence interval (CI) were calculated to assess diagnostic performance using Real-Q assay as reference method. The degree of concordance between Exdia Ag and Real-Q assays was tested by Cohen’s kappa coefficient (κ). The median values of Ct value were also calculated and compared using the Wilcoxon rank-sum test. The SPSS statistical package version 24.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analyses. The significance level was set at P ≤ 0.05.
Between December 2021, and March 2022, a total of 2,534 paired samples were used for analysis. Among them, excluding 11 samples (six samples showed indeterminate results with Real-Q and five samples showed invalid results with Exdia Ag assay), the diagnostic performance was assessed excluding these samples.
Among the 2,523 samples, 149 showed positive results and 2,374 showed negative results with Real-Q assay. Compared to the Real-Q assay, the overall sensitivity and specificity of the Exdia Ag assay was 77.2% (115/149) (95% CI, 69.6 - 83.7) and 99.8% (2,370/2,374) (95% CI, 99.6 - 99.9), respectively. The accuracy was 98.5% (2,485/2,523) (95% CI, 99.6 - 99.9) and κ value was 0.85 (95% CI, 0.80 - 0.90) with almost perfect agreement.
The PPV and NPV in this study cohort, with prevalence of 5.9% (149/2,523), were 96.6% (95% CI, 91.5 - 98.7) and 98.6% (95% CI, 98.1 - 98.9), respectively. Because PPV and NPV vary according to the prevalence, with prevalence of 20% and 0.5%; the results obtained were 99.1% and 69.7%, respectively, for PPV, and 94.6% and 99.9%, respectively, for NPV.
The distribution of Exdia Ag assay result according to Ct value of Real-Q assay (RdRp gene) is shown in Table 1. The sensitivity of Exdia Ag assay was highest in samples with low Ct values of Real-Q assay, with a sensitivity of 98.3% for Ct ≤ 15, decreased slightly to 92.7% for 15 < Ct ≤ 20 dropped further to 72.2% for 20 < Ct ≤ 25 and was lowest (only 13.3%) for 30 < Ct.
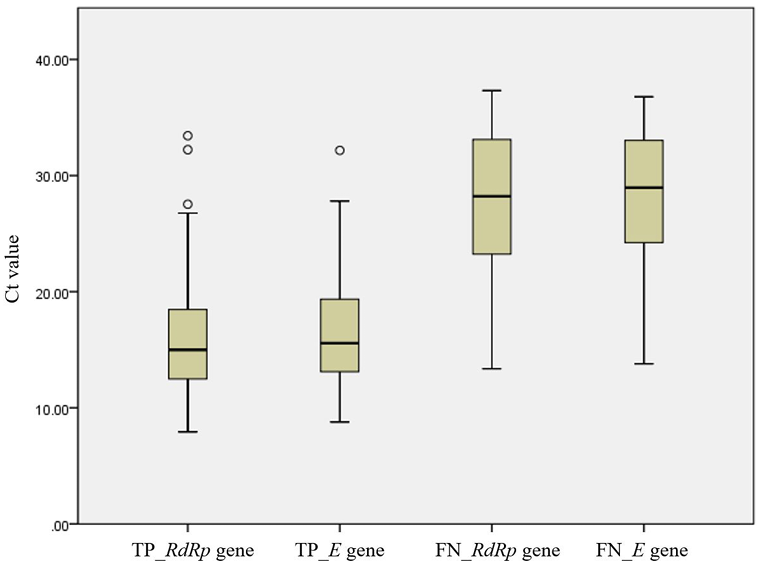
For 115 concordant Exdia Ag-positive/Real-Q-positive results, the median Ct value for RdRp and envelope (E) genes (interquartile range, IQR) of Real-Q results was 15.0 (12.4 - 18.6) and 15.6 (13.1 - 19.5), respectively. For 34 discordant Exdia Ag-negative/Real-Q-positive results, the median Ct value for RdRp and E gene (IQR) of Real-Q results was 28.2 (22.8 - 33.2) and 29.0 (23.8 - 33.1), respectively. We observed a significantly lower Ct value for concordant Exdia Ag-positive/Real-Q-positive samples than that in discordant Exdia Ag-negative/Real-Q-positive samples (p< 0.0001, both RdRp and E gene) (Fig. 1).
Some reports evaluating the performance of rapid antigen assays exhibited wide range of sensitivity (41.0%-80.6%) depending on the study design (e.g., patient population) and sample types [4,7,8]. Additionally, we observed only four false positive cases (0.2%) for Exdia assay, consistent with other previous studies (about 1.0% or less false positive rate for antigen assays) [9]. Researchers mentioned that those false positive results of antigen assays are not associated with any specific respiratory pathogen [6,10].
Although the sensitivity of Exdia Ag assay became lower as the Ct value of RdRp gene became higher, the majority (134/149, 89.9%) of rRT-PCR positive patients showed Ct value of ≤ 30 (Table 1). This is in line with a large meta-analysis review reported that overall cumulative sensitivity was 95.8% and 96.5% for Ct value ≤ 25 and Ct value ≤ 20, respectively [11]. In similar, we observed that the Exdia Ag assay showed 92.4% and 96.0% sensitivity for samples with Ct value ≤ 25 and ≤ 20, respectively. According to the preliminary evaluation of the performance of other rapid antigen kit, it also showed 66.7% and 90.0% sensitivity for samples with Ct value ≤ 25 and ≤ 20, respectively (Data not shown). Because we used frozen residual nasopharyngeal swab specimens for this pilot study, the possibility that the sensitivity was affected by frozen specimens cannot be ruled out [12]. Considering that minimal RNA genome copy number of 106 genome copies per ml of specimen, corresponding to a Ct value ≤ 25, represents the amount of infectious virus particles required for successful virus propagation in cell culture [13-15] rapid antigen test kit will be useful for rapid detection and triage of patients in ED.
This study has a strength in that we included a large sample size for which both tests were performed simultaneously. However, this study has several limitations. First, we did not consider the proportion of asymptomatic patients in our cohort due to absence of clinical information. In addition, it was not possible to conduct a sensitivity analysis based on the presence or absence of symptoms. Second, as two different samples were collected for performing each assay, this may have influenced the evaluation results.
We conclude that the Exdia assay shows its strength in high specificity (99.8%) and moderately sensitive (92.4%) in patients with high viral loads (Ct ≤ 25), but supplementary test should be performed for patients with the negative test results especially before admission into the hospital to prevent missed spread of SARSCoV-2.
Ethics statement
This study was approved by the Institutional Review Board of Seoul St. Mary’s Hospital (No. KC22RASI0213).
Acknowledgements
We thank the entire laboratory staff for their efforts engaged in the performance of rapid and accurate tests during the COVID-19 pandemic.
REFERENCES
1. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25.
.
2. Hong KH, Kim GJ, Roh KH, Sung H, Lee J, Kim SY, et al. Update of guidelines for laboratory diagnosis of COVID-19 in Korea. Ann Lab Med 2022;42:391-7.
.
3. Diel R and Nienhaus A. Point-of-care COVID-19 antigen testing in German emergency rooms - a cost-benefit analysis. Pulmonology 2021;28:164-72.
.
4. Caruana G, Croxatto A, Kampouri E, Kritikos A, Opota O, Foerster M, et al. Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 2021;9:798.
.
5. Leli C, Di Matteo L, Gotta F, Cornaglia E, Vay D, Megna I, et al. Performance of a SARSCoV-2 antigen rapid immunoassay in patients admitted to the emergency department. Int J Infect Dis 2021;110:135-40.
.
6. Möckel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, et al. SARSCoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 2021;26:213-20.
.
7. Greub G, Caruana G, Schweitzer M, Imperiali M, Muigg V, Risch M, et al. Multicenter technical validation of 30 rapid antigen tests for the detection of SARS-CoV-2 (VALIDATE). Microorganisms 2021;9:2589.
.
8. Kritikos A, Caruana G, Brouillet R, Miroz JP, Abed-Maillard S, Stieger G, et al. Sensitivity of rapid antigen testing and RT-PCR performed on nasopharyngeal swabs versus saliva samples in COVID-19 hospitalized patients: results of a prospective comparative trial (RESTART). Microorganisms 2021;9:1910.
.
9. Routsias JG, Mavrouli M, Tsoplou P, Dioikitopoulou K, Tsakris A. Diagnostic performance of rapid antigen tests (RATs) for SARS-CoV-2 and their efficacy in monitoring the infectiousness of COVID-19 patients. Sci Rep 2021;11:22863.
.
10. Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a singlecentre laboratory evaluation study. Lancet Microbe 2021;2:e311-9.
.
11. Brümmer LE, Katzenschlager S, Gaeddert M, Erdmann C, Schmitz S, Bota M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: a living systematic review and metaanalysis. PLoS Med 2021;18:e1003735.
.
12. Kweon OJ, Lim YK, Kim HR, Choi Y, Kim MC, Choi SH, et al. Evaluation of rapid SARS-CoV-2 antigen tests, AFIAS COVID-19 Ag and ichroma COVID-19 Ag, with serial nasopharyngeal specimens from COVID-19 patients. PLoS One 2021;16:e0249972.
.
13. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465-9.
.
14. Kim MC, Cui C, Shin KR, Bae JY, Kweon OJ, Lee MK, et al. Duration of culturable SARSCoV-2 in hospitalized patients with Covid-19. N Engl J Med 2021;384:671-3.
.
15. van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021;12:267.
.
Table 1
Distribution of Exdia Ag test result according to cycle threshold (Ct) values of Real-Q assay (RdRp gene)
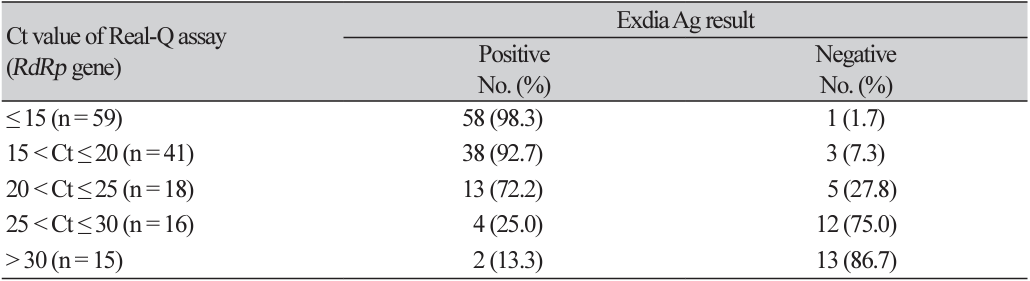
Fig. 1.
Box-plots of cycle threshold (Ct) values distributions according to the concordance results between Exdia Ag and Real-Q assays. Solid lines in box-plots represent the Ct value medians, box-plots and whiskers represent the Ct values distribution. Abbreviations: TP, Exdia Ag-positive/Real-Q-positive samples; FN, Exdia Ag-negative/Real-Qpositive samples; RdRp, RNA-dependent RNA polymerase gene; E, envelope gene; Exdia Ag, Exdia COVID-19 antigen; Real-Q, Real-Q 2019-nCoV Detection KIT.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download