1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424.

2. Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA. 2006; 295:2164–7.

3. Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013; 2013:965212.

4. Mora-Guzman I, Munoz de Nova JL, Marin-Campos C, et al. Efficiency of the Bethesda System for Thyroid Cytopathology. Cir Esp (Engl Ed). 2018; 96:363–8.

5. Vaccarella S, Franceschi S, Bray F, Wild CP, Plummer M, Dal Maso L. Worldwide thyroid-cancer epidemic? The increasing impact of overdiagnosis. N Engl J Med. 2016; 375:614–7.

6. Daskalakis A, Kostopoulos S, Spyridonos P, et al. Design of a multiclassifier system for discriminating benign from malignant thyroid nodules using routinely H&E-stained cytological images. Comput Biol Med. 2008; 38:196–203.

7. Chain K, Legesse T, Heath JE, Staats PN. Digital image-assisted quantitative nuclear analysis improves diagnostic accuracy of thyroid fine-needle aspiration cytology. Cancer Cytopathol. 2019; 127:501–13.

8. Gerhard R, Teixeira S, Gaspar da Rocha A, Schmitt F. Thyroid fineneedle aspiration cytology: is there a place to virtual cytology? Diagn Cytopathol. 2013; 41:793–8.

9. Fragopoulos C, Pouliakis A, Meristoudis C, et al. Radial basis function artificial neural network for the investigation of thyroid cytological lesions. J Thyroid Res. 2020; 2020:5464787.

10. Schmitt WR. Punção aspirativa por agulha fina e a sua importância diagnóstica nas lesões de tireoide [Fine needle aspiration and its diagnostic importance in thyroid lesions]. Porto: Universidade do Porto;2011.
11. Shurbaji MS, Gupta PK, Frost JK. Nuclear grooves: a useful criterion in the cytopathologic diagnosis of papillary thyroid carcinoma. Diagn Cytopathol. 1988; 4:91–4.

12. Ali SZ, Cibas ES. The Bethesda System for Reporting Thyroid Cytopathology. Cham: Springer;2018.
13. Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid. 2017; 27:1341–6.

14. Ali SZ, VanderLaan PA. The Bethesda System for Reporting Thyroid Cytopathology. Cham: Springer;2023.
15. LiVolsi VA. Papillary thyroid carcinoma: an update. Mod Pathol. 2011; 24 Suppl 2:S1–9.

16. Baloch ZW, LiVolsi VA, Asa SL, et al. Diagnostic terminology and morphologic criteria for cytologic diagnosis of thyroid lesions: a synopsis of the National Cancer Institute Thyroid Fine-Needle Aspiration State of the Science Conference. Diagn Cytopathol. 2008; 36:425–37.

17. Batistatou A, Scopa CD. Pathogenesis and diagnostic significance of nuclear grooves in thyroid and other sites. Int J Surg Pathol. 2009; 17:107–10.

18. Francis IM, Das DK, Sheikh ZA, Sharma PN, Gupta SK. Role of nuclear grooves in the diagnosis of papillary thyroid carcinoma: a quantitative assessment on fine needle aspiration smears. Acta Cytol. 1995; 39:409–15.
19. Gould E, Watzak L, Chamizo W, Albores-Saavedra J. Nuclear grooves in cytologic preparations: a study of the utility of this feature in the diagnosis of papillary carcinoma. Acta Cytol. 1989; 33:16–20.
20. Das DK. Intranuclear cytoplasmic inclusions in fine-needle aspiration smears of papillary thyroid carcinoma: a study of its morphological forms, association with nuclear grooves, and mode of formation. Diagn Cytopathol. 2005; 32:264–8.

21. Yang YJ, Demirci SS. Evaluating the diagnostic significance of nuclear grooves in thyroid fine needle aspirates with a semiquantitative approach. Acta Cytol. 2003; 47:563–70.
22. Rupp M, Ehya H. Nuclear grooves in the aspiration cytology of papillary carcinoma of the thyroid. Acta Cytol. 1989; 33:21–6.
23. Scopa CD, Melachrinou M, Saradopoulou C, Merino MJ. The significance of the grooved nucleus in thyroid lesions. Mod Pathol. 1993; 6:691–4.
24. Silverman JF, Frable WJ. The use of the diff-quik stain in the immediate interpretation of fine-needle aspiration biopsies. Diagn Cytopathol. 1990; 6:366–9.

25. Dey P. Basic and advanced laboratory techniques in histopathology and cytology. Singapore: Springer Singapore;2018.
26. Bhambhani S, Kashyap V, Das DK. Nuclear grooves. Valuable diagnostic feature in May-Grunwald-Giemsa-stained fine needle aspirates of papillary carcinoma of the thyroid. Acta Cytol. 1990; 34:809–12.
27. Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019; 20:e253–61.

28. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology: new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019; 16:703–15.

29. Pouliakis A, Karakitsou E, Margari N, et al. Artificial neural networks as decision support tools in cytopathology: past, present, and future. Biomed Eng Comput Biol. 2016; 7:1–18.

30. Gupta N, Sarkar C, Singh R, Karak AK. Evaluation of diagnostic efficiency of computerized image analysis based quantitative nuclear parameters in papillary and follicular thyroid tumors using paraffin-embedded tissue sections. Pathol Oncol Res. 2001; 7:46–55.

31. Valentim FO, Coelho BP, Miot HA, et al. Follicular thyroid lesions: is there a discriminatory potential in the computerized nuclear analysis? Endocr Connect. 2018; 7:907–13.

32. Yashaswini R, Suresh TN, Sagayaraj A. Cytological evaluation of thyroid lesions by nuclear morphology and nuclear morphometry. J Cytol. 2017; 34:197–202.

33. Karakitsos P, Cochand-Priollet B, Guillausseau PJ, Pouliakis A. Potential of the back propagation neural network in the morphologic examination of thyroid lesions. Anal Quant Cytol Histol. 1996; 18:494–500.
34. Ramos HE, Vale J, Lopes S, et al. Nuclear score evaluation in follicular-patterned thyroid lesions using optical and digital environments. Endocrine. 2022; 77:486–92.

35. Kezlarian B, Lin O. Artificial intelligence in thyroid fine needle aspiration biopsies. Acta Cytol. 2021; 65:324–9.

36. Legesse T, Parker L, Heath J, Staats PN. Distinguishing non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) from classic and invasive follicular-variant papillary thyroid carcinomas based on cytologic features. J Am Soc Cytopathol. 2019; 8:11–7.

37. Kuzan TY, Guzelbey B, Turan Guzel N, Kuzan BN, Cakir MS, Canbey C. Analysis of intra-observer and inter-observer variability of pathologists for non-benign thyroid fine needle aspiration cytology according to Bethesda system categories. Diagn Cytopathol. 2021; 49:850–5.

38. Cibas ES, Baloch ZW, Fellegara G, et al. A prospective assessment defining the limitations of thyroid nodule pathologic evaluation. Ann Intern Med. 2013; 159:325–32.

39. Thompson LD, Poller DN, Kakudo K, Burchette R, Nikiforov YE, Seethala RR. An international interobserver variability reporting of the nuclear scoring criteria to diagnose noninvasive follicular thyroid neoplasm with papillary-like nuclear features: a validation study. Endocr Pathol. 2018; 29:242–9.

40. Liu Z, Bychkov A, Jung CK, et al. Interobserver and intraobserver variation in the morphological evaluation of noninvasive follicular thyroid neoplasm with papillary-like nuclear features in Asian practice. Pathol Int. 2019; 69:202–10.

41. House JC, Henderson-Jackson EB, Johnson JO, et al. Diagnostic digital cytopathology: are we ready yet? J Pathol Inform. 2013; 4:28.

42. Vodovnik A. Diagnostic time in digital pathology: a comparative study on 400 cases. J Pathol Inform. 2016; 7:4.

43. Jiang P, Ergu D, Liu F, Cai Y, Ma B. A review of Yolo algorithm developments. Procedia Comput Sci. 2022; 199:1066–73.

44. Sanyal P, Mukherjee T, Barui S, Das A, Gangopadhyay P. Artificial intelligence in cytopathology: a neural network to identify papillary carcinoma on thyroid fine-needle aspiration cytology smears. J Pathol Inform. 2018; 9:43.

45. Guan Q, Wang Y, Ping B, et al. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: a pilot study. J Cancer. 2019; 10:4876–82.

46. Duan W, Gao L, Liu J, et al. Computer-assisted fine-needle aspiration cytology of thyroid using two-stage refined convolutional neural network. Electronics. 2022; 11:4089.

47. Nguyen DUC, Lee YM, Park J. An Ensemble deep learning for automatic prediction of papillary thyroid carcinoma using fine needle aspiration cytology. Expert Syst Appl. 2021; 188:115927.
48. Aloqaily A, Polonia A, Campelos S, et al. Digital versus optical diagnosis of follicular patterned thyroid lesions. Head Neck Pathol. 2021; 15:537–43.

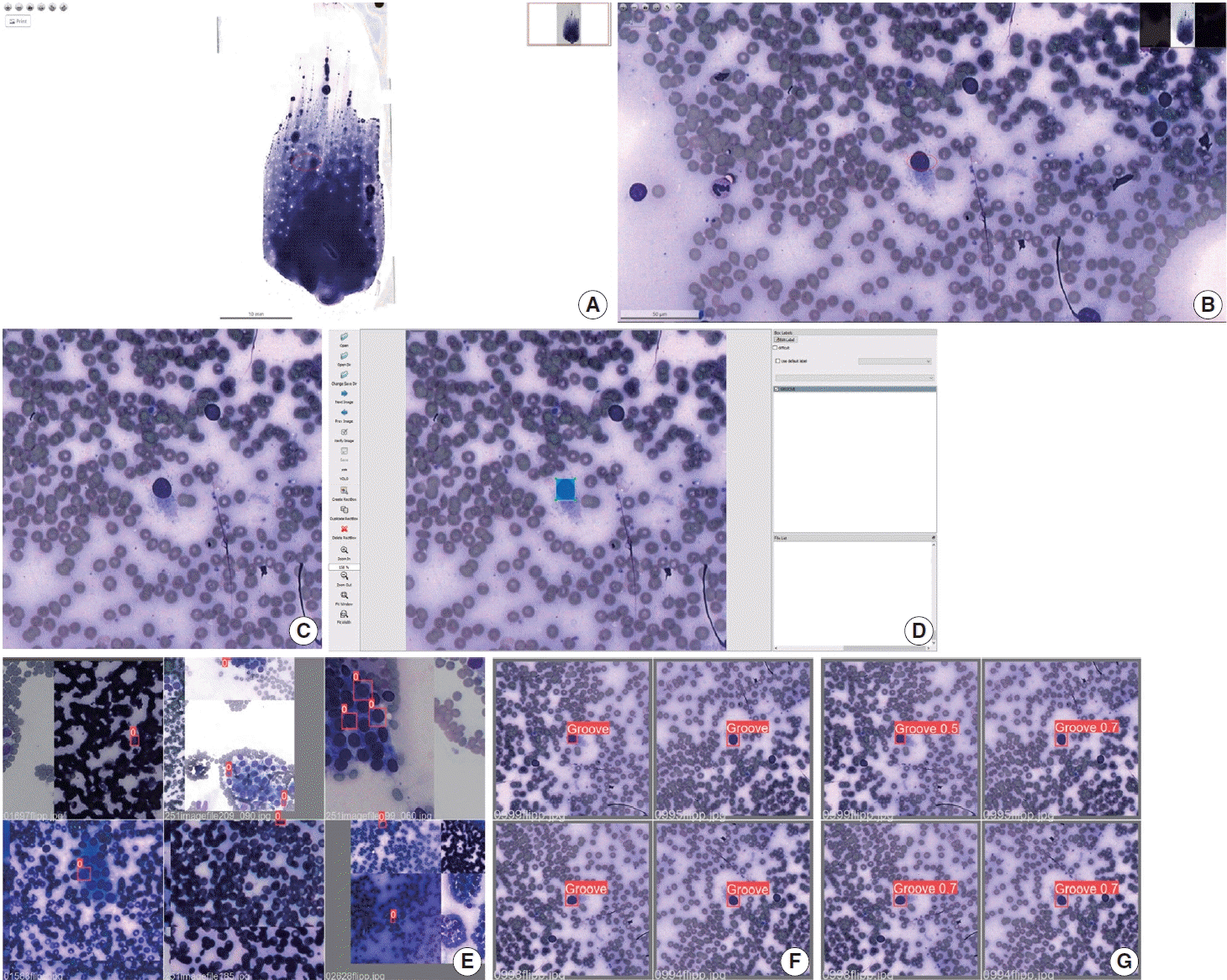
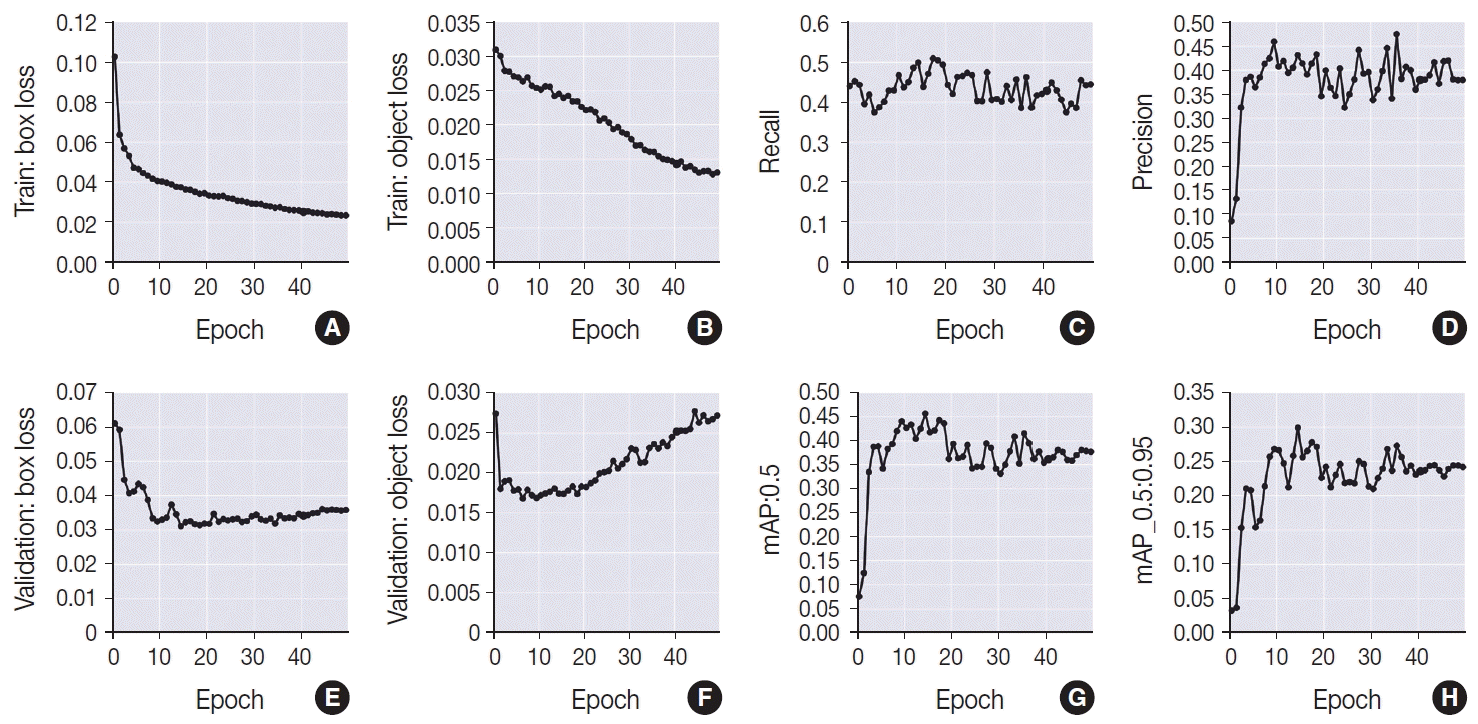




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download