Dear Editor,
Low-density lipoprotein–cholesterol (LDL-C) levels strongly correlate with cardiovascular disease risk. Guidelines recommend targeting low LDL-C levels to reduce this risk. Therefore, precise measurement is essential [1].
The LDL-C reference method requires ultracentrifugation by trained personnel and large serum samples [2, 3]. Equations, such as Friedewald, Martin–Hopkins, or Sampson, are often used instead to calculate LDL-C levels [4, 5]. Equations have limitations, particularly for the calculation LDL-C≤70 mg/dL or triglycerides levels (TG) ≥400 mg/dL. Furthermore, bias and imprecision from the different measurements used in the calculation can adversely affect LDL-C calculation accuracy [6].
LDL-C clinical decision limits are based on the Friedewald equation. Despite the additional cost, the direct LDL-C method (dLDL-C) is faster than ultracentrifugation and more accurate than equations. dLDL-C results may vary either among manufacturers or depending on the reagent-calibrator-instrument combination. Therefore, it is advisable to compare the dLDL-C with the reference method [7].
We assessed and compared the dLDL-C and equations for calculating LDL-C against the reference method to identify quick and accurate alternatives. This was a prospective study developed in Spain, from March 2022 to March 2023.
In total, 212 serum samples stored at 4ºC were analyzed and categorized into two groups (IRB; PI-21-036): group 1 (TG≥400 mg/dL, positive lipemic index, N=113) and group 2 (TG≤200 mg/dL, LDL-C≤70 mg/dL, N=99). Group 1 was subdivided into 1a (TG=400–500 mg/dL, N=62) and 1b (TG=500–900 mg/dL, N=51). Group 2 was subdivided into 2a (LDL-C=10–40 mg/dL, N=49) and 2b (LDL-C=40–70 mg/dL, N=50).
dLDL-C level was measured using the enzymatic selective protection method [8] with an AU5800 analyzer (Beckman Coulter, Brea, CA, USA). The dLDL-C calibrator value is traceable to the reference method. LDL-C was calculated using the Friedewald, Martin–Hopkins (180-cell strata), and Sampson equations. Very-low-density lipoprotein–cholesterol (VLDL-C) was isolated from 1.5 mL of serum utilizing sequential density gradient ultracentrifugation at 100,000×g for 18 hrs with an F50L fixed-angle rotor (Thermo Scientific, Basingstoke, UK) using KBr for density adjustment (1,006 g/mL). TC, HDL-C, TG, and VLDL-C contents were measured using the AU5800 analyzer, and LDL-C level was calculated as follows: LDL-C=TC−HDL-C−VLDL-C [2, 3, 4].
Statistical analysis was conducted using MedCalc v19.6 (MedCalc Software, Ostend, Belgium). Means were compared using t-test for paired samples to compare the different methods against the reference method.
Bias between methods was calculated as follows:
where Cn and Cx represent the LDL-C determined using the reference and alternative methods, respectively. Bias was compared with the reference change value (RCV), which was calculated for LDL-C considering a unilateral Z statistic with 95% confidence (Z=1.65), as follows:
where CVa and Cvi are the analytical CV and within-subject biological variation according to the European Federation of Clinical Chemistry and Laboratory Medicine Biological Variation Database, respectively [9]. The RCV was calculated to be 20.0% based on CVa=2.18 and CVi=8.3. Bland–Altman difference plots were used to compare the reference and alternative methods in groups 1 and 2.
Table 1 reveals significant differences in LDL-C between the reference and alternative methods, except for dLDL-C in group 1. However, the bias did not exceed the RCV for dLDL-C and Martin–Hopkins. In group 2, LDL-C differed significantly from those determined using the reference method for all alternative methods; however, the bias did not exceed the RCV for dLDL-C.
In groups 1a and 1b, LDL-C measured using the reference method differed significantly from the values determined using the alternative methods, except for dLDL-C. The bias never exceeded the RCV for dLDL-C in groups 1a and 1b nor for Martin–Hopkins in group 1a. In groups 2a and 2b, all methods differ significantly from the reference method; however, the bias did not exceed the RCV for dLDL-C (Table 1).
Bland–Altman plots demonstrated minimal bias of dLDL-C vs. the reference method in groups 1 and 2 (Fig. 1).
Notably, for TG≥400 mg/dL, dLDL-C was the superior method. For TG=500–900 mg/dL, results from all alternative methods differed from ultracentrifugation results, except for dLDL-C. For TG=400–500 mg/dL, both dLDL-C and Martin–Hopkins methods were suitable, with dLDL-C being the most accurate. dLDL-C is applicable for LDL-C<70 mg/dL.
For LDL-C<70 mg/dL or TG≥400 mg/dL, Friedewald, Martin–Hopkins, and Sampson tend to underestimate LDL-C. Martin–Hopkins and Sampson equations reportedly have improved accuracy over Friedewald with TG≥400 mg/dL or LDL-C<70 mg/dL [5, 10]. However, these studies did not compare these methods with ultracentrifugation, but with dLDL-C.
In conclusion, the study showed that both equations and the Friedewald method continue to underestimate LDL-C. The findings support that dLDL-C is an excellent choice for TG≥400 mg/dL or LDL-C<70 mg/dL. These results are specific for the Beckman Coulter dLDL-C; differences among manufacturers have been reported [7]. Similarly, TC, HDL-C, and TG assays used to calculate LDL-C are not perfectly standardized among manufacturers.
Notes
AUTHOR CONTRIBUTIONS
Martínez-Bujidos M, Morales-Indiano C and Fernández-Prendes C contributed to the conception and design of the study; Rodríguez-Domínguez J, Piedra-Aguilera A, and Fernández-Prendes C interpreted the results; Piedra-Aguilera A performed the statistical analysis; Rodríguez-Domínguez J, Piedra-Aguilera A, Malumbres-Serrano S and Fernández-Prendes C drafted the manuscript; and Fernández-Prendes C supervised the study. All authors read and approved the final manuscript.
References
1. Wilson PWF, Polonsky TS, Miedema MD, Khera A, Kosinski AS, Kuvin JT. 2019; Systematic review for the 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 139:e1144–61. DOI: 10.1161/CIR.0000000000000626.

2. Havel RJ, Eder HA, Bragdon JH. 1955; The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 34:1345–53. DOI: 10.1172/JCI103182. PMID: 13252080. PMCID: PMC438705.

3. Ordovas JM. 1998; Fast Ultracentrifugation Methods for the Separation of Plasma Lipoproteins. Methods Mold Biol. 110:93–103. DOI: 10.1385/1-59259-582-0:93. PMID: 9918041.

4. Frost PH, Havel RJ. 1998; Rationale for use of non-high-density lipoprotein cholesterol rather than low-density lipoprotein cholesterol as a tool for lipoprotein cholesterol screening and assessment of risk and therapy. Am J Cardiol. 81:26B–31B. DOI: 10.1016/S0002-9149(98)00034-4. PMID: 9526810.

5. Ertürk Zararsız G, Bolat S, Cephe A, Kochan N, Yerlitaş SI, Doğan HO, Zararsız G. 2022; Validation of Friedewald, Martin-Hopkins and Sampson low-density lipoprotein cholesterol equations. PLoS One. 17:e0263860. DOI: 10.1371/journal.pone.0263860. PMID: 35559957. PMCID: PMC9106156. PMID: aa397892c5b8434a940f35a94e3ae0ed.

6. Martin SS, Blaha MJ, Elshazly MB, Brinton EA, Toth PP, McEvoy JW, et al. 2013; Friedewald-estimated versus directly measured low-density lipoprotein cholesterol and treatment implications. J Am Coll Cardiol. 62:732–9. DOI: 10.1016/j.jacc.2013.01.079. PMID: 23524048.

7. Nauck M, Warnick GR, Rifai N. 2002; Methods for measurement of LDL-cholesterol: a critical assessment of direct measurement by homogeneous assays versus calculation. Clin Chem. 48:236–54. DOI: 10.1093/clinchem/48.2.236. PMID: 11805004.

8. Miki Y. 1999; A homogeneous assay for the selective measurement of LDL-cholesterol in serum. Enzymatic selective protection method. Clin Lab. 45:398–401.
9. EFLM Biological Variation Database. https://biologicalvariation.eu. Updated on April 2023.
10. Azimi V, Farnsworth CW, Roper SM. 2022; Comparison of the Friedewald equation with Martin and Sampson equations for estimating LDL cholesterol in hypertriglyceridemic adults. Clin Biochem. 108:1–4. DOI: 10.1016/j.clinbiochem.2022.07.005. PMID: 35905970.

Fig. 1
Bland–Altman plots for the alternative methods and the ultracentrifugation (reference) method. (A) dLDL-C, (B) Friedewald, (C) Martin–Hopkins, and (D) Sampson methods applied in group 1. (E) dLDL-C, (F) Friedewald, (G) Martin–Hopkins, and (H) Sampson methods applied in group 2.
Abbreviations: TG, triglycerides; LDL-C, low-density lipoprotein–cholesterol; dLDL-C, direct low-density lipoprotein–cholesterol.
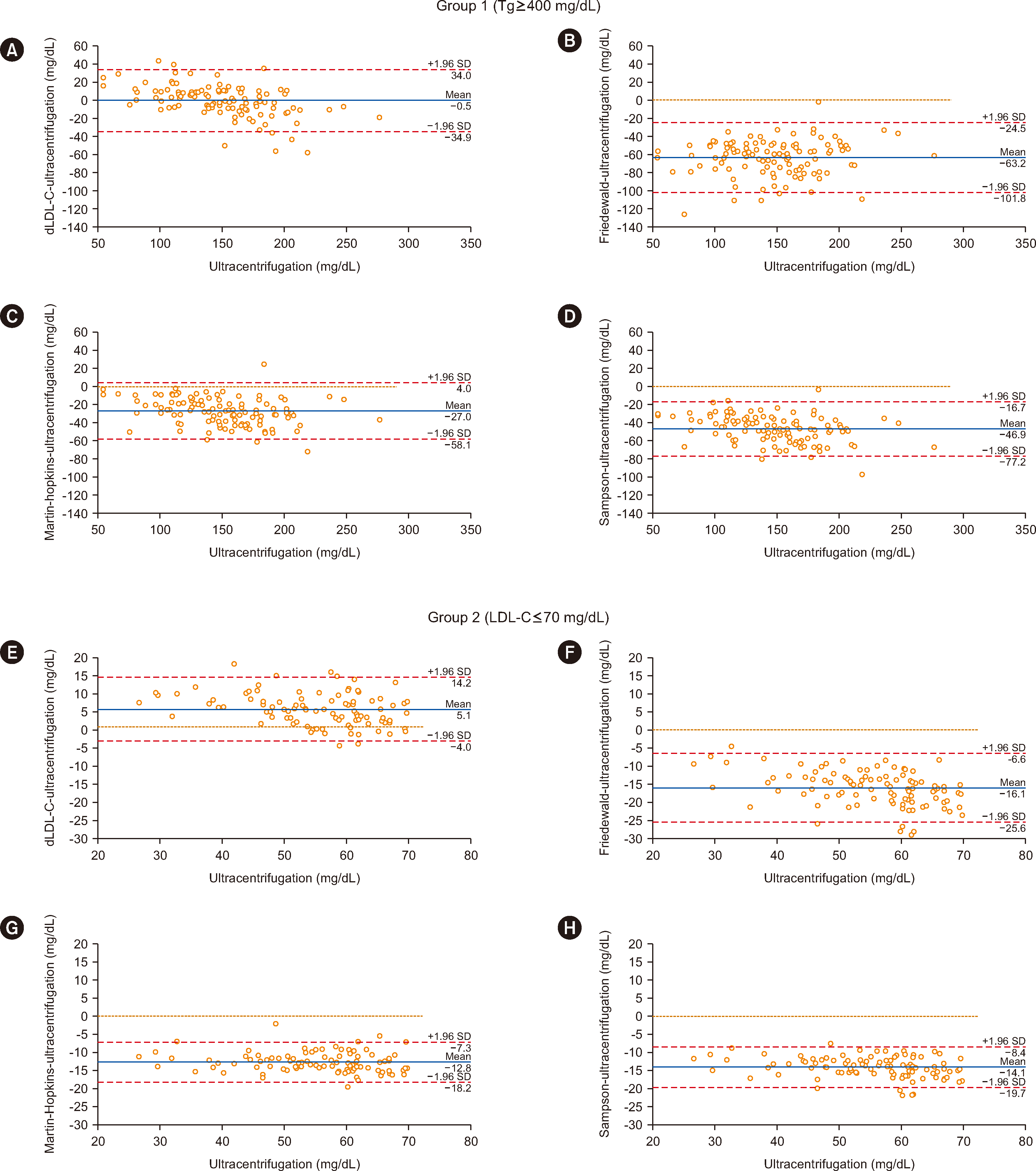
Table 1
Comparison of methods for direct LDL-C assay and estimated equations for LDL-C with a reference method
| Group | Variable | Ultracentrifugation* | dLDL-C | Friedewald | Martin–Hopkins | Sampson |
|---|---|---|---|---|---|---|
| Group 1 (TG≥400 mg/dL) | Median (IQR) | 148 (116–177) | 149 (130–170) | 85 (59–112) | 121 (101–142) | 101 (81–122) |
| P | 0.7745 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | 0.5 | −42.4 | −18.5 | −31.7 | ||
| Group 1a (TG=400–500 mg/dL) | Median (IQR) | 151 (115–180) | 149 (122–174) | 93 (61–124) | 126 (101–151) | 106 (78–132) |
| P | 0.4070 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | −1.5 | −38.4 | −16.6 | −30.1 | ||
| Group 1b (TG=500–900 mg/dL) | Median (IQR) | 148 (124–174) | 148 (132–165) | 77 (55–98) | 114 (102–134) | 98 (82–112) |
| P | 0.8201 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | −0.2 | −48.3 | −22.7 | −34.0 | ||
| Group 2 (LDL-C≤70 mg/dL) | Median (IQR) | 58 (49–62) | 61 (54–68) | 41 (34–46) | 45 (37–50) | 43 (36–48) |
| P | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | 5.3 | −29.6 | −22.9 | −25.4 | ||
| Group 2a (LDL-C=10–40 mg/dL) | Median (IQR) | 49 (44–54) | 55 (51–63) | 34 (29–38) | 36 (30–41) | 36 (29–40) |
| P | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | 10.9 | −31.8 | −25.8 | −26.7 | ||
| Group 2b (LDL-C=40–70 mg/dL) | Median (IQR) | 62 (58–66) | 65 (61–69) | 46 (43–49) | 49 (46–53) | 48 (45–51) |
| P | <0.0001 | <0.0001 | <0.0001 | <0.0001 | ||
| Bias (%) | 5.8 | −25.1 | −20.2 | −21.9 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download