Abstract
Purpose
This study aims to determine the association between pre- and postoperative radiotherapy (PORT) circulating tumor DNA (ctDNA) dynamics and oncological outcomes in patients with residual triple-negative breast cancer who underwent surgery after neoadjuvant chemotherapy (NAC).
Materials and Methods
Between March 2019 and July 2020, 11 nonmetastatic patients with residual disease who underwent surgery after NAC were prospectively enrolled. In each patient, tumor specimens obtained during surgery and blood samples collected at three time points during PORT (T0: pre-PORT, T1: 3 weeks after PORT, T2: 1 month after PORT) were sequenced, targeting 38 cancer-related genes. Disease-free survival (DFS) was evaluated and the association between DFS and ctDNA dynamics was analyzed.
Results
At T0, ctDNA was detected in three (27.2%) patients. The ctDNA dynamics were as follows: two showed a decreasing ctDNA variant allele frequency (VAF) and reached zero VAF at T2, while one patient exhibited an increasing VAF during PORT and maintained an elevated VAF at T2. After a median follow-up of 48 months, two patients experienced distant metastasis without any locoregional failures. All failures occurred in patients with ctDNA positivity at T0 and a decreased VAF after PORT. The 4-year DFS rates according to the T0 ctDNA status were 67% (positive ctDNA) and 100% (negative ctDNA) (p=0.032).
Conclusion
More than a quarter of the patients with residual disease after post-NAC surgery exhibited pre-PORT ctDNA positivity, and ctDNA positivity was associated with poor DFS. For patients with pre-PORT ctDNA positivity, the administration of a more effective systemic treatment should be considered.
Breast cancer is one of the most commonly diagnosed malignancies in women, and its incidence is increasing [1]. The treatment of many types of cancer largely depends on the histology and disease stage. In breast cancer, subtypes based on immunohistochemistry are also associated with prognosis and are important for determining treatment strategies [2]. Efforts have been made to further classify the disease and personalize the treatment for patients with breast cancer. Recent advancements in liquid biopsy techniques have enabled the development of novel approaches for cancer diagnosis and treatment. Liquid biopsy uses peripheral blood instead of directly obtaining tumor tissue, making it less invasive and allowing easy sample collection throughout the disease course [3]. Circulating tumor DNA (ctDNA), which is cell-free DNA originating from tumor cells, can be analyzed using liquid biopsy. ctDNAs can be used to detect minimal residual disease (MRD), which is associated with poor prognosis in some cancer types. Tracking MRD may enable the tailoring of treatment based on recurrence risk, thereby potentially avoiding adverse events in patients with favorable prognosis and improving treatment outcomes in patients with unfavorable prognosis [4].
In patients with locally advanced breast cancer, neoadjuvant chemotherapy (NAC) is administered prior to surgery [5]. Achieving complete response after NAC is associated with improved prognosis, particularly in the triple-negative subtype [6]. Adjuvant capecitabine is indicated for therapeutic intensification in patients with residual tumors after NAC, as this indicates poor survival [7]. Further risk stratification may be possible using ctDNAs. Assessing ctDNA levels after definitive treatment may provide additional information on MRD, allowing for the prediction of poor prognosis and adjustment of treatment decisions. We conducted a prospective observational study to analyze ctDNA dynamics before and after postoperative radiation therapy (PORT) [8]. In this report, we present additional follow-up data for the patient cohort because intriguing results were observed during the follow-up period.
The details of the methodology are described in a previously published article [8], and a brief description is included in this report. Between March 2019 and July 2020, eleven triple-negative breast cancer patients who underwent NAC, surgery, and PORT were enrolled. All patients exhibited macroscopic residual disease in their surgical specimens. Anthracycline was used in 10 patients (90.9%) and taxane was used in all patients as the NAC regimen. The median time from NAC completion to surgery was 25 days (range, 20 to 32 days). PORT was initiated 4-5 weeks after surgery, and the PORT field encompassed the breast or chest wall and the regional lymph node area. All except one patient (90.9%) started the administration of adjuvant capecitabine 3-4 weeks after completion of PORT. Blood samples for ctDNA analysis were collected before PORT (T0), 3 weeks after PORT initiation (T1), and 1 month after PORT completion (T2). The median time from surgery to the first blood sampling was 32 days (range, 23 to 39 days). Ultra-deep sequencing (average depth 5,884.8× for cell-free DNA and 3,993.4× for genomic DNA) was performed on tumor tissues, three plasma samples, and leukocytes from each patient. Customized baits targeting 38 cancer-related genes were used (LiquidSCAN v2-PanCancer panel, GENINUS, Seoul, Korea). The variant allele frequency (VAF) of ctDNA was measured and changes at different time points were evaluated. A schematic diagram of the study process is shown in Fig. 1.
Additional follow-up data were retrieved from the patients’ medical records. Actuarial rates of disease-free survival (DFS), measured from the initiation of NAC, were calculated using the Kaplan-Meier method. The event for DFS was defined as any disease progression. Log-rank p-values were calculated for patients with and without ctDNA. The characteristics, ctDNA dynamics, and clinical course of patients who experienced recurrence are described. Statistical significance was set at p < 0.05. All statistical analyses were conducted using the R software ver. 4.2.1 (R Foundation for Statistical Computing, Vienna, Austria).
The median follow-up period was 48 months (range, 36 to 54 months). At T0, ctDNA was detected in three patients (27.2%). Detailed ctDNA detection results have been described in previous publication [8]. Two patients (P1 and P8) with detectable ctDNA levels at T0 experienced recurrence. The actuarial DFS rates according to ctDNA detection are illustrated in Fig. 2. A statistically significant difference in DFS was observed between patients with and without ctDNA (p=0.032). The 4-year DFS rate was 67% for patients with detectable ctDNA at T0 and 100% for those without detectable ctDNA. Patient characteristics and treatment specifics are summarized in Table 1. VAF in the plasma of these patients was the highest at T0 and later decreased, reaching zero at T2. Both patients with recurrence initially had advanced disease that spread to the internal mammary and/or supraclavicular lymph nodes. One patient (P9) had increased plasma VAF at T2, but no recurrence was reported at 48 months of follow-up.
One patient (P1) with recurrence had distant metastasis to the bone and pleura 33 months after NAC initiation. Palliative radiotherapy and chemotherapy were administered to the involved bone. However, brain metastasis progressed. The patient received intrathecal methotrexate, talazoparib, and whole-brain radiotherapy, although further disease progression was later reported. Another patient (P8) with a recurrence had distant metastases. Multiple liver, bone, and lymphangitic metastases were detected 51 months after NAC initiation. The patient received palliative chemotherapy.
In a previous report, we hypothesized that the decrease in ctDNA VAF during and after PORT could be interpreted as the eradication of localized MRD by radiotherapy [8]. However, on further follow-up, both patients with decreasing ctDNA VAF levels experienced distant recurrence. In contrast, patients with increased ctDNA levels after PORT did not experience tumor recurrence. This result differs slightly from our initial expectations. The detection of ctDNA appears to be a strong predictive factor for recurrence. However, decreased ctDNA levels during and after PORT may not be a prognostic factor.
The detection of ctDNA and its prognostic significance in NAC for breast cancer has been the subject of several studies. Several studies have shown that ctDNA levels are significantly associated with the treatment outcomes. However, the exact time points for ctDNA detection suggested in these studies differed. Studies have demonstrated the prognostic significance of ctDNA detection at baseline [9], during or after NAC [10-13], and after surgery [14]. The prognostic significance of ctDNA detection was also observed in this study, as two out of three patients with detectable ctDNA in postoperative blood sampling experienced recurrence, whereas no recurrence was reported in patients without ctDNA. The results of this study contribute to the evidence support the prognostic significance of postoperative ctDNA detection in patients with breast cancer treated with NAC.
In contrast to the prognostic significance of ctDNA detection, the pre- and post-radiotherapy ctDNA dynamics observed in this study did not match our initial assumptions. Previous studies have shown that a decrease in ctDNA is associated with a better tumor response and treatment outcomes in various cancer types treated with chemotherapy, including breast cancer [12,15,16]. In radiotherapy, the dynamics of circulating viral DNA in cancers related to viral etiology have been observed in several studies. Better treatment outcomes were associated with the clearance of circulating viral DNA in patients who underwent definitive radiotherapy for human papillomavirus-related cervical cancer and Epstein-Barr virus-related nasopharyngeal cancer [17,18]. Post-radiotherapy ctDNA detection has also been associated with poor prognosis in patients who undergo definitive radiotherapy for esophageal cancer [19]. Based on these previous reports, we assumed that a decrease in ctDNA VAF may indicate MRD eradication and, therefore, correlate with better outcomes. However, in this study, patients with ctDNA detected after surgery and those with ctDNA clearance after PORT experienced recurrence. Differences in neoadjuvant, definitive, and adjuvant treatment settings may have caused this phenomenon. In neoadjuvant and definitive treatments, the clinically visible tumor burden remains, but in adjuvant settings, most of the tumor burden is removed. Therefore, the clearance of ctDNA after neoadjuvant and definitive treatments may have stronger prognostic implications than a decrease in ctDNA levels after PORT. Another possible explanation is that PORT may effectively eradicate locoregional MRD but have limited effects on distant metastasis. Both patients had distant metastases throughout their clinical course and no locoregional recurrence was reported. MRD occurring in the locoregional area may have been eradicated by PORT, leading to a decrease in ctDNA VAF. It should be noted that both patients had internal mammary and supraclavicular lymph node metastases, which made complete surgical removal challenging. However, this assumption cannot be validated solely based on the observations of the present study.
In this study, one patient (P9) had an increased ctDNA VAF at the post–radiation therapy (RT) 1-month follow-up, but did not experience recurrence. Although this patient was receiving adjuvant capecitabine when blood was sampled, this change in ctDNA could not be explained as chemotherapy-related. Some studies have observed a transient peak in ctDNA during chemotherapy; however, this peak usually appeared a few days after the initiation of chemotherapy [20,21]. In this patient, the post-RT 1-month blood sampling was conducted three weeks after the initiation of adjuvant capecitabine. The absence of recurrence in this patient may have been due to chance.
While showing the prognostic significance of ctDNA detection, this study also demonstrated the discrepancies between tumor and ctDNA mutation profiles. In this study, the same 38 cancer-related genes were observed in tumor and blood samples, and the variants observed in ctDNA did not appear in tumors. The exact methodology for ctDNA detection varies among studies. Some studies utilized personalized or ‘bespoke’ ctDNA. This approach selects mutations from each tumor mutation profile, and ctDNA detection is performed for these selected mutations only. The detection of ctDNA using this methodology showed good correlation with treatment outcomes [10,11,22]. However, assuming discrepancies between tumor and ctDNA mutation profiles, such as those shown in this study, patient-specific ctDNA detection may have significant false negatives. One possible explanation for the discrepancies between tumor and ctDNA mutation profiles is tumor heterogeneity. Several studies reported tumor heterogeneity in breast cancer and its association of poor prognosis [23,24]. Due to spatial heterogeneity, samples obtained from specific sites of surgical tumor specimens may not contain mutations in metastatic clones. Researchers have observed differences in mutation profiles between tumors and ctDNA, and have suggested that analyzing ctDNA can help overcome tumor heterogeneity [25-27]. An optimal approach for ctDNA detection that considers intrapatient heterogeneity should be addressed in future research.
ctDNA testing may provide genotyping information that can aid the selection of appropriate targeted agents. The PlasmaMATCH trial allocated patients to different treatment arms with targeted agents based on ctDNA testing results, and the use of targeted therapies against ERBB2 and AKT1 mutations showed promising clinical feasibility [28]. In this study, ctDNA analysis revealed the presence of EGFR, CTNNB1, and MAP2K4 mutations. However, clinical implications of these mutations remain unclear. While EGFR overexpression is often observed in triple-negative breast cancer, EGFR mutations are rare [29]. Unlike for lung adenocarcinoma, there are currently no approved targeted agents for breast cancer with EGFR mutations. Additionally, there are no available agents that specifically target CTNNB1 and MAP2K4 mutations. Although ctDNA analysis has the potential to identify druggable targets in breast cancer, careful selection of target genes for sequencing is crucial for achieving this purpose.
Immunotherapy is actively used for the treatment of TNBC. The addition of pembrolizumab to cytotoxic chemotherapy has shown promising results for both operable and inoperable/metastatic TNBC [30,31]. ctDNA detection can also be integrated into immunotherapy. A recent trial, c-TRAK TN, aimed to assess the feasibility of ctDNA surveillance and the commencement of pembrolizumab immunotherapy. However, this trial failed to demonstrate the effectiveness of ctDNA-guided pembrolizumab, presumably because of the small number of patients who underwent treatment [32]. Other trials are also investigating the feasibility of integrating ctDNA detection and immunotherapy, such as the PERSEVERE trial (NCT04849364). Further data on this approach will be available in the future.
A major limitation of this study is the small number of patients enrolled. Although a significant difference in DFS was observed between patients with ctDNA detection and those without ctDNA detection, the interaction of other prognostic factors, such as initial disease extent, should be analyzed. Notably, both patients who had a recurrence in this study also had more advanced disease that metastasized to the internal mammary and/or supraclavicular lymph nodes at the initial clinical presentation. The small cohort size limited further statistical analyses of the independent association between ctDNA and treatment outcomes. The detection sensitivity of ctDNA in this study may have been limited by the analysis of only 38 genes. The observations from this study were hypothesis-generating rather than confirmative, and further studies are needed for clarification. Despite these limitations, ctDNA dynamics and their association with treatment outcomes in PORT settings for breast cancer have rarely been studied. This report provides valuable information for these settings.
In conclusion, the results of this prospective study, after 4 years of follow-up, showed that ctDNA detection during PORT for breast cancer has prognostic significance. However, contrary to our initial assumptions, decreasing ctDNA levels during and after PORT did not necessarily indicate better treatment outcomes. Therefore, in the setting of PORT for breast cancer, ctDNA detection may have stronger prognostic implications than ctDNA dynamics. Nonetheless, as patients with ctDNA and recurrence did not experience locoregional recurrence, an explanation for localized MRD eradication with PORT is still plausible.
Notes
Ethical Statement
This study was approved by the Institutional Review Board of Samsung Medical Center prior to study initiation (approval number 2018-10-137). Written informed consent was obtained from each participant before enrollment.
ACKNOWLEDGMENTS
This study was supported by grants from the Ministry of Science and ICT, Republic of Korea (NRF-2019R1F1A1062069) and Samsung Medical Center (SMO 1200821 and SMO 1200391).
References
1. Kang MJ, Won YJ, Lee JJ, Jung KW, Kim HJ, Kong HJ, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2019. Cancer Res Treat. 2022; 54:330–44.

2. Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010; 7:e1000279.

3. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic: implementation issues and future challenges. Nat Rev Clin Oncol. 2021; 18:297–312.

4. Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020; 1:276–90.

5. Korde LA, Somerfield MR, Carey LA, Crews JR, Denduluri N, Hwang ES, et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J Clin Oncol. 2021; 39:1485–505.

6. Spring LM, Fell G, Arfe A, Sharma C, Greenup R, Reynolds KL, et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res. 2020; 26:2838–48.

7. Masuda N, Lee SJ, Ohtani S, Im YH, Lee ES, Yokota I, et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N Engl J Med. 2017; 376:2147–59.

8. Kim H, Kim YJ, Park D, Park WY, Choi DH, Park W, et al. Dynamics of circulating tumor DNA during postoperative radiotherapy in patients with residual triple-negative breast cancer following neoadjuvant chemotherapy: a prospective observational study. Breast Cancer Res Treat. 2021; 189:167–75.

9. Li S, Lai H, Liu J, Liu Y, Jin L, Li Y, et al. Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol. 2020; 4:244–57.

10. Cavallone L, Aguilar-Mahecha A, Lafleur J, Brousse S, Aldamry M, Roseshter T, et al. Prognostic and predictive value of circulating tumor DNA during neoadjuvant chemotherapy for triple negative breast cancer. Sci Rep. 2020; 10:14704.

11. Ortolan E, Appierto V, Silvestri M, Miceli R, Veneroni S, Folli S, et al. Blood-based genomics of triple-negative breast cancer progression in patients treated with neoadjuvant chemotherapy. ESMO Open. 2021; 6:100086.
12. Magbanua MJ, Swigart LB, Wu HT, Hirst GL, Yau C, Wolf DM, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol. 2021; 32:229–39.

13. Cailleux F, Agostinetto E, Lambertini M, Rothe F, Wu HT, Balcioglu M, et al. Circulating tumor DNA after neoadjuvant chemotherapy in breast cancer is associated with disease relapse. JCO Precis Oncol. 2022; 6:e2200148.

14. Radovich M, Jiang G, Hancock BA, Chitambar C, Nanda R, Falkson C, et al. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 2020; 6:1410–5.

15. Moser T, Waldispuehl-Geigl J, Belic J, Weber S, Zhou Q, Hasenleithner SO, et al. On-treatment measurements of circulating tumor DNA during FOLFOX therapy in patients with colorectal cancer. NPJ Precis Oncol. 2020; 4:30.

16. Song Y, Hu C, Xie Z, Wu L, Zhu Z, Rao C, et al. Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl Lung Cancer Res. 2020; 9:269–79.

17. Han K, Leung E, Barbera L, Barnes E, Croke J, Di Grappa MA, et al. Circulating human papillomavirus DNA as a biomarker of response in patients with locally advanced cervical cancer treated with definitive chemoradiation. JCO Precis Oncol. 2018; 2:1–8.

18. He SS, Wang Y, Bao Y, Cai XY, Yang XL, Chen DM, et al. Dynamic changes in plasma Epstein-Barr virus DNA load during treatment have prognostic value in nasopharyngeal carcinoma: a retrospective study. Cancer Med. 2018; 7:1110–7.

19. Jia R, Zhao CH, Li PS, Liu RR, Zhang Y, Chen HE, et al. Postradiation circulating tumor DNA as a prognostic factor in locally advanced esophageal squamous cell carcinoma. Oncol Lett. 2021; 21:68.

20. Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015; 26:1715–22.

21. Kruger S, Heinemann V, Ross C, Diehl F, Nagel D, Ormanns S, et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann Oncol. 2018; 29:2348–55.

22. Bratman SV, Yang SY, Iafolla MA, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020; 1:873–81.

23. McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019; 26:2191–9.

24. Ma F, Guan Y, Yi Z, Chang L, Li Q, Chen S, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2020; 146:1359–68.

25. Gao J, Wang H, Zang W, Li B, Rao G, Li L, et al. Circulating tumor DNA functions as an alternative for tissue to overcome tumor heterogeneity in advanced gastric cancer. Cancer Sci. 2017; 108:1881–7.
26. Perdigones N, Murtaza M. Capturing tumor heterogeneity and clonal evolution in solid cancers using circulating tumor DNA analysis. Pharmacol Ther. 2017; 174:22–6.

27. Fridland S, Choi J, Nam M, Schellenberg SJ, Kim E, Lee G, et al. Assessing tumor heterogeneity: integrating tissue and circulating tumor DNA (ctDNA) analysis in the era of immunooncology: blood TMB is not the same as tissue TMB. J Immunother Cancer. 2021; 9:e002551.
28. Turner NC, Kingston B, Kilburn LS, Kernaghan S, Wardley AM, Macpherson IR, et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 2020; 21:1296–308.

29. Kim A, Jang MH, Lee SJ, Bae YK. Mutations of the epidermal growth factor receptor gene in triple-negative breast cancer. J Breast Cancer. 2017; 20:150–9.

30. Schmid P, Cortes J, Pusztai L, McArthur H, Kummel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020; 382:810–21.

31. Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im SA, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020; 396:1817–28.
32. Turner NC, Swift C, Jenkins B, Kilburn L, Coakley M, Beaney M, et al. Results of the c-TRAK TN trial: a clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann Oncol. 2023; 34:200–11.

Fig. 1.
Schematic diagram for process of this study. ctDNA, circulating tumor DNA; RT, radiation therapy.
Fig. 2.
Actuarial rates of disease-free survival according to detection of circulating tumor DNA (ctDNA).
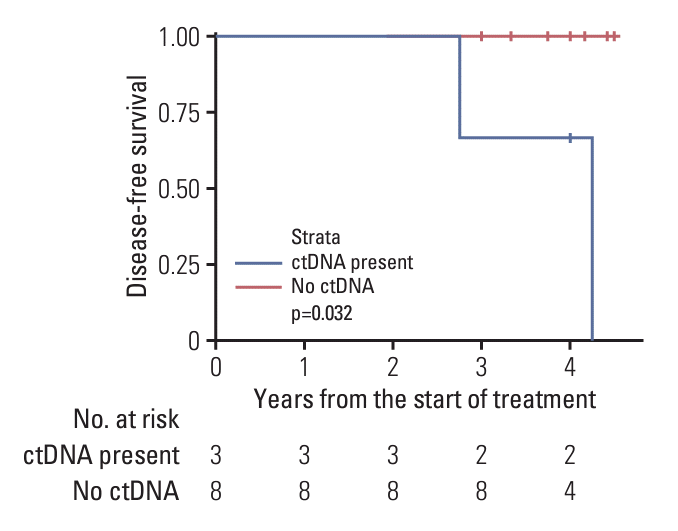
Table 1.
Patient characteristics and treatment




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download