1. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013; 19:1423–37.

2. Ren X, Kang B, Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol. 2018; 19:211.

3. Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009; 9:302–12.

4. Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell. 2020; 37:471–84.

5. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020; 15:1484–506.

6. Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020; 17:159–62.

7. Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single-cell technologies for monitoring immune systems. Nat Immunol. 2014; 15:128–35.

8. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017; 171:1611–24.

9. Young MD, Mitchell TJ, Vieira Braga FA, Tran MG, Stewart BJ, Ferdinand JR, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018; 361:594–9.

10. Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023; 41:404–20.

11. Zhuang X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat Methods. 2021; 18:18–22.

12. Tan WCC, Nerurkar SN, Cai HY, Ng HH, Wu D, Wee YT, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun (Lond). 2020; 40:135–53.

13. Hickey JW, Neumann EK, Radtke AJ, Camarillo JM, Beuschel RT, Albanese A, et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat Methods. 2022; 19:284–95.

14. Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015; 16:57–66.

15. Allam M, Cai S, Coskun AF. Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics. NPJ Precis Oncol. 2020; 4:11.

16. Wang N, Li X, Wang R, Ding Z. Spatial transcriptomics and proteomics technologies for deconvoluting the tumor microenvironment. Biotechnol J. 2021; 16:e2100041.

17. Maniatis S, Petrescu J, Phatnani H. Spatially resolved transcriptomics and its applications in cancer. Curr Opin Genet Dev. 2021; 66:70–7.

18. Zheng B, Fang L. Spatially resolved transcriptomics provide a new method for cancer research. J Exp Clin Cancer Res. 2022; 41:179.

19. Bassiouni R, Gibbs LD, Craig DW, Carpten JD, McEachron TA. Applicability of spatial transcriptional profiling to cancer research. Mol Cell. 2021; 81:1631–9.

20. Marx V. Method of the Year: spatially resolved transcriptomics. Nat Methods. 2021; 18:9–14.

21. Lewis SM, Asselin-Labat ML, Nguyen Q, Berthelet J, Tan X, Wimmer VC, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021; 18:997–1012.

22. Rao A, Barkley D, Franca GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021; 596:211–20.

23. Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021; 22:627–44.

24. McGinnis LM, Ibarra-Lopez V, Rost S, Ziai J. Clinical and research applications of multiplexed immunohistochemistry and in situ hybridization. J Pathol. 2021; 254:405–17.
25. Ahmed R, Augustine R, Valera E, Ganguli A, Mesaeli N, Ahmad IS, et al. Spatial mapping of cancer tissues by OMICS technologies. Biochim Biophys Acta Rev Cancer. 2022; 1877:188663.

26. Kuczkiewicz-Siemion O, Sokol K, Puton B, Borkowska A, Szumera-Cieckiewicz A. The role of pathology-based methods in qualitative and quantitative approaches to cancer immunotherapy. Cancers (Basel). 2022; 14:3833.

27. Akhoundova D, Rubin MA. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell. 2022; 40:920–38.

28. Wu Y, Cheng Y, Wang X, Fan J, Gao Q. Spatial omics: Navigating to the golden era of cancer research. Clin Transl Med. 2022; 12:e696.

29. Ahmed R, Zaman T, Chowdhury F, Mraiche F, Tariq M, Ahmad IS, et al. Single-cell RNA sequencing with spatial transcriptomics of cancer tissues. Int J Mol Sci. 2022; 23:3042.

30. Walker BL, Cang Z, Ren H, Bourgain-Chang E, Nie Q. Deciphering tissue structure and function using spatial transcriptomics. Commun Biol. 2022; 5:220.

31. Moses L, Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022; 19:534–46.

32. Asp M, Bergenstrahle J, Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays. 2020; 42:e1900221.

33. Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016; 353:78–82.

34. Gracia Villacampa E, Larsson L, Mirzazadeh R, Kvastad L, Andersson A, Mollbrink A, et al. Genome-wide spatial expression profiling in formalin-fixed tissues. Cell Genom. 2021; 1:100065.

35. Harouaka RA, Nisic M, Zheng SY. Circulating tumor cell enrichment based on physical properties. J Lab Autom. 2013; 18:455–68.

36. Nagasawa S, Kuze Y, Maeda I, Kojima Y, Motoyoshi A, Onishi T, et al. Genomic profiling reveals heterogeneous populations of ductal carcinoma in situ of the breast. Commun Biol. 2021; 4:438.

37. Chu Y, Dai E, Li Y, Han G, Pei G, Ingram DR, et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat Med. 2023; 29:1550–62.

38. Zollinger DR, Lingle SE, Sorg K, Beechem JM, Merritt CR. GeoMx RNA assay: high multiplex, digital, spatial analysis of RNA in FFPE tissue. Methods Mol Biol. 2020; 2148:331–45.

39. Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol. 2020; 38:586–99.

40. Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011; 11:46.

41. Vadakekolathu J, Minden MD, Hood T, Church SE, Reeder S, Altmann H, et al. Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci Transl Med. 2020; 12:eaaz0463.

42. Keane C, Law SC, Gould C, Birch S, Sabdia MB, Merida de Long L, et al. LAG3: a novel immune checkpoint expressed by multiple lymphocyte subsets in diffuse large B-cell lymphoma. Blood Adv. 2020; 4:1367–77.

43. Stewart RL, Matynia AP, Factor RE, Varley KE. Spatially-resolved quantification of proteins in triple negative breast cancers reveals differences in the immune microenvironment associated with prognosis. Sci Rep. 2020; 10:6598.

44. Hwang WL, Jagadeesh KA, Guo JA, Hoffman HI, Yadollahpour P, Reeves JW, et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat Genet. 2022; 54:1178–91.

45. Danaher P, Kim Y, Nelson B, Griswold M, Yang Z, Piazza E, et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. 2022; 13:385.

46. Pich-Bavastro C, Yerly L, Di Domizio J, Tissot-Renaud S, Gilliet M, Kuonen F. Activin A-mediated polarization of cancerassociated fibroblasts and macrophages confers resistance to checkpoint immunotherapy in Skin Cancer. Clin Cancer Res. 2023; 29:3498–513.
47. Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014; 11:360–1.

48. Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016; 92:342–57.

49. Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging: spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015; 348:aaa6090.
50. He S, Bhatt R, Brown C, Brown EA, Buhr DL, Chantranuvatana K, et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat Biotechnol. 2022; 40:1794–806.

51. Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015; 10:442–58.

52. Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wahlby C, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013; 10:857–60.

53. Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018; 361:eaat5691.

54. Chen F, Tillberg PW, Boyden ES. Optical imaging: expansion microscopy. Science. 2015; 347:543–8.
55. Xia C, Fan J, Emanuel G, Hao J, Zhuang X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci U S A. 2019; 116:19490–9.

56. Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc Natl Acad Sci U S A. 2016; 113:14456–61.

57. Janesick AJ, Shelansky R, Gottscho A, Wagner F, Rouault M, Beliakoff G, et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue. Preprint at bioRxiv:
https://doi.org/10.1101/2022.10.06.510405 (2022).

58. Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol. 2016; 1:aaf6925.

59. Akturk G, Sweeney R, Remark R, Merad M, Gnjatic S. Multiplexed Immunohistochemical Consecutive Staining on Single Slide (MICSSS): multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol Biol. 2020; 2055:497–519.

60. Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife. 2018; 7:e31657.

61. Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014; 20:436–42.

62. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014; 11:417–22.

63. Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018; 174:968–81.

64. Schurch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020; 182:1341–59.

65. Ahmed FS, Gaule P, McGuire J, Patel K, Blenman K, Pusztai L, et al. PD-L1 protein expression on both tumor cells and macrophages are associated with response to neoadjuvant durvalumab with chemotherapy in triple-negative breast cancer. Clin Cancer Res. 2020; 26:5456–61.

66. Na HY, Park Y, Nam SK, Koh J, Kwak Y, Ahn SH, et al. Prognostic significance of natural killer cell-associated markers in gastric cancer: quantitative analysis using multiplex immunohistochemistry. J Transl Med. 2021; 19:529.

67. Park Y, Seo AN, Koh J, Nam SK, Kwak Y, Ahn SH, et al. Expression of the immune checkpoint receptors PD-1, LAG3, and TIM3 in the immune context of stage II and III gastric cancer by using single and chromogenic multiplex immunohistochemistry. Oncoimmunology. 2021; 10:1954761.

68. Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017; 19:203–17.

69. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019; 35:588–602.

70. Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Methods Enzymol. 2020; 635:1–20.

71. Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013; 110:11982–7.

72. Li C, Ma H, Wang Y, Cao Z, Graves-Deal R, Powell AE, et al. Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer. J Clin Invest. 2014; 124:2172–87.

73. Gerdes MJ, Gokmen-Polar Y, Sui Y, Pang AS, LaPlante N, Harris AL, et al. Single-cell heterogeneity in ductal carcinoma in situ of breast. Mod Pathol. 2018; 31:406–17.

74. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014; 70:46–58.

75. Halse H, Colebatch AJ, Petrone P, Henderson MA, Mills JK, Snow H, et al. Multiplex immunohistochemistry accurately defines the immune context of metastatic melanoma. Sci Rep. 2018; 8:11158.

76. Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017; 8:15095.

77. Guo M, Yuan F, Qi F, Sun J, Rao Q, Zhao Z, et al. Expression and clinical significance of LAG-3, FGL1, PD-L1 and CD8(+) T cells in hepatocellular carcinoma using multiplex quantitative analysis. J Transl Med. 2020; 18:306.
78. Majonis D, Herrera I, Ornatsky O, Schulze M, Lou X, Soleimani M, et al. Synthesis of a functional metal-chelating polymer and steps toward quantitative mass cytometry bioassays. Anal Chem. 2010; 82:8961–9.

79. Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, et al. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl. 2007; 46:6111–4.

80. Han G, Spitzer MH, Bendall SC, Fantl WJ, Nolan GP. Metalisotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat Protoc. 2018; 13:2121–48.

81. Rana R, Gomez-Biagi RF, Bassan J, Nitz M. Signal amplification for imaging mass cytometry. Bioconjug Chem. 2019; 30:2805–10.

82. Wang YJ, Traum D, Schug J, Gao L, Liu C, Consortium H, et al. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metab. 2019; 29:769–83.

83. Damond N, Engler S, Zanotelli VR, Schapiro D, Wasserfall CH, Kusmartseva I, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019; 29:755–68.

84. Keren L, Bosse M, Thompson S, Risom T, Vijayaragavan K, McCaffrey E, et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv. 2019; 5:eaax5851.

85. Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018; 174:1373–87.

86. Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022; 14:68.

87. Kim TY, Lee HS, Bae J, Park Y, Oh DY, Yoon J, et al. Analysis of the role of tumor microenvironment on clinical outcomes of chemotherapy in patients with advanced gastric cancer using high plex digital spatial profiling. J Clin Oncol. 2023; 41(4 Suppl):456.

88. Zhao T, Chiang ZD, Morriss JW, LaFave LM, Murray EM, Del Priore I, et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022; 601:85–91.

89. Schulz D, Zanotelli VR, Fischer JR, Schapiro D, Engler S, Lun XK, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018; 6:531.

90. Hoch T, Schulz D, Eling N, Gomez JM, Levesque MP, Bodenmiller B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci Immunol. 2022; 7:eabk1692.

91. Jiang S, Chan CN, Rovira-Clave X, Chen H, Bai Y, Zhu B, et al. Combined protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments. Immunity. 2022; 55:1118–34.

92. Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021; 4:5.

93. Fu Y, Jung AW, Torne RV, Gonzalez S, Vohringer H, Shmatko A, et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer. 2020; 1:800–10.

94. Petukhov V, Xu RJ, Soldatov RA, Cadinu P, Khodosevich K, Moffitt JR, et al. Cell segmentation in imaging-based spatial transcriptomics. Nat Biotechnol. 2022; 40:345–54.

95. Zhao Y, Wang K, Hu G. DIST: spatial transcriptomics enhancement using deep learning. Brief Bioinform. 2023; 24:bbad013.

96. Xu C, Jin X, Wei S, Wang P, Luo M, Xu Z, et al. DeepST: identifying spatial domains in spatial transcriptomics by deep learning. Nucleic Acids Res. 2022; 50:e131.

97. Liu T, Fang ZY, Li X, Zhang LN, Cao DS, Yin MZ. Graph deep learning enabled spatial domains identification for spatial transcriptomics. Brief Bioinform. 2023; 24:bbad146.

98. Lee RY, Ng CW, Rajapakse MP, Ang N, Yeong JP, Lau MC. The promise and challenge of spatial omics in dissecting tumour microenvironment and the role of AI. Front Oncol. 2023; 13:1172314.

99. Pham D, Tan X, Balderson B, Xu J, Grice LF, et al. Robust mapping of spatiotemporal trajectories and cell–cell interactions in healthy and diseased tissues. Nat Commun. 2023; 14:7739.

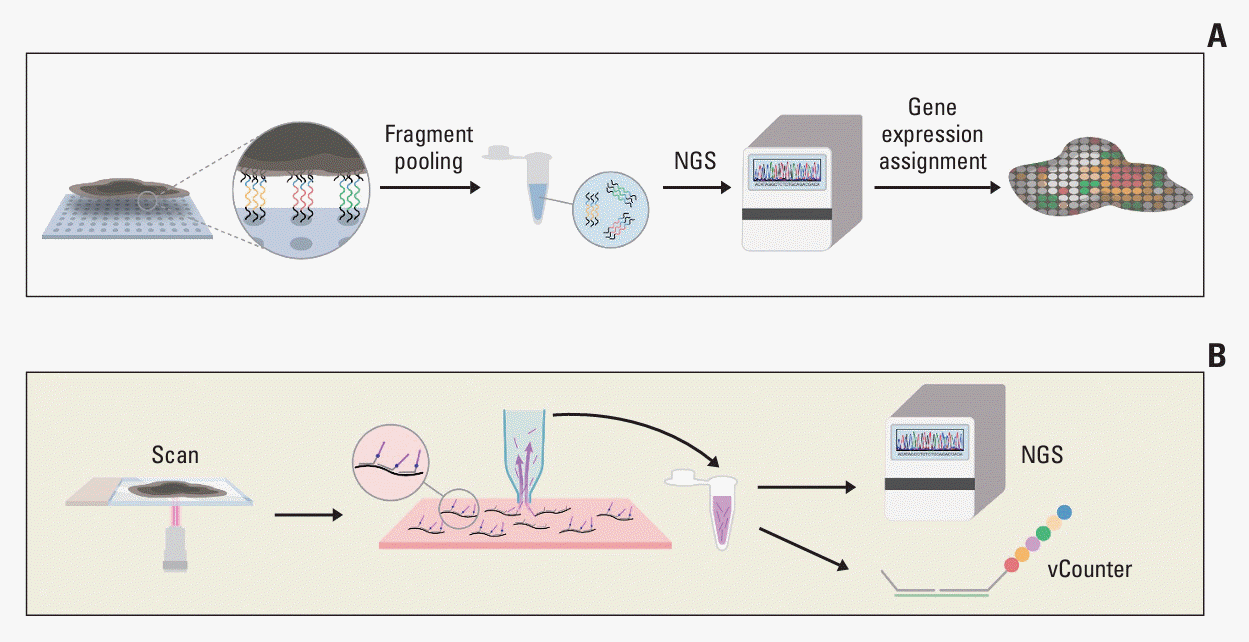
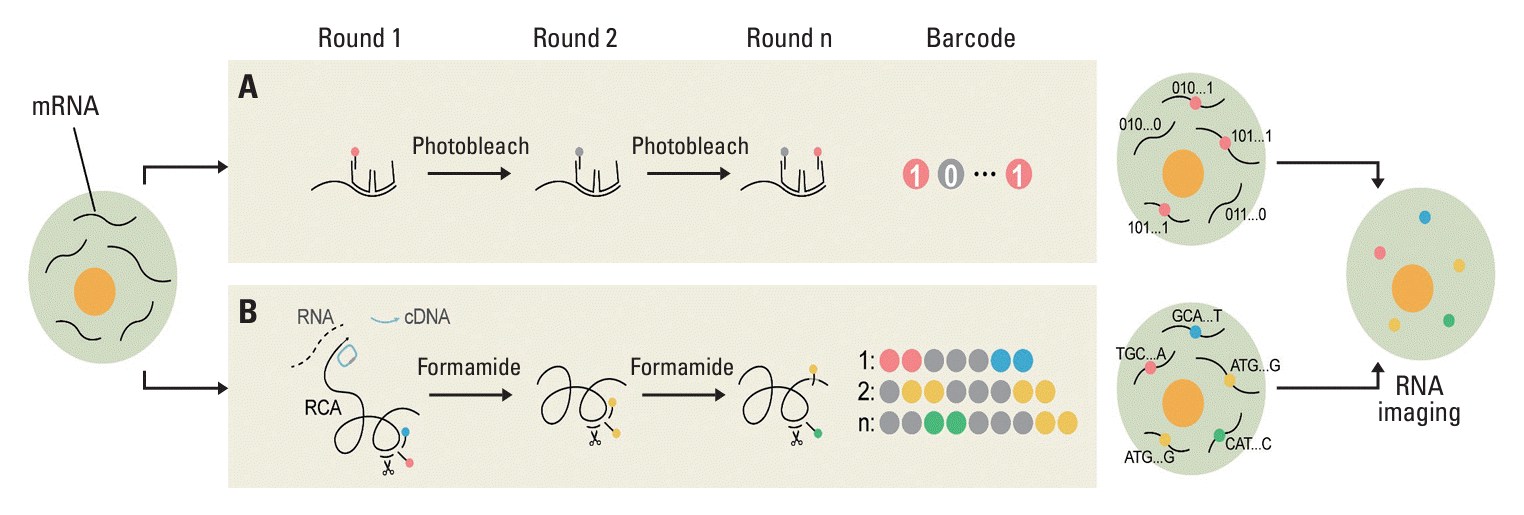
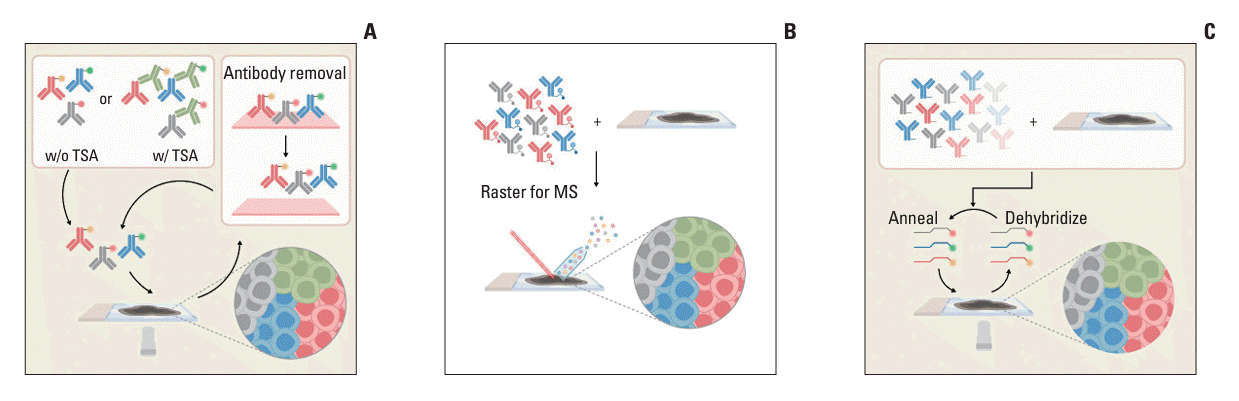




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download