1. Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021; 397:1750–69.

2. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–48.

3. Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019; 25:4255–63.

4. Garcia-Murillas I, Chopra N, Comino-Mendez I, Beaney M, Tovey H, Cutts RJ, et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019; 5:1473–8.

5. Parsons HA, Rhoades J, Reed SC, Gydush G, Ram P, Exman P, et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin Cancer Res. 2020; 26:2556–64.

6. Lipsyc-Sharf M, de Bruin EC, Santos K, McEwen R, Stetson D, Patel A, et al. Circulating tumor DNA and late recurrence in high-risk hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer. J Clin Oncol. 2022; 40:2408–19.

7. Li S, Lai H, Liu J, Liu Y, Jin L, Li Y, et al. Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol. 2020; 4:PO.19.00292.

8. ClinicalTrials.gov. A trial using ctDNA blood tests to detect cancer cells after standard treatment to trigger additional treatment in early stage triple negative breast cancer patients (c-TRAK-TN) [Internet]. Bethesda, MD: National Library of Medicine;2017. [cited 2023 Dec 3]. Available from:
https://clinicaltrials.gov/ct2/show/NCT03145961.
9. ClinicalTrials.gov. A prospective, phase II trial using ctDNA to initiate post-operation boost therapy after adjuvant chemotherapy in TNBC (Artemis) [Internet]. Bethesda, MD: National Library of Medicine;2017. cited 2023 Dec 3]. Available from:
https://clinicaltrials.gov/ct2/show/NCT04803539.
10. ClinicalTrials.gov. A prospective, phase II trial using ctDNA to initiate post-operation boost therapy after NAC in TNBC (Apollo) [Internet]. Bethesda, MD: National Library of Medicine;2020. [cited 2023 Dec 3]. Available from:
https://clinicaltrials.gov/ct2/show/NCT04501523.
11. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016; 534:47–54.
12. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016; 7:11479.
13. Lang GT, Jiang YZ, Shi JX, Yang F, Li XG, Pei YC, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. 2020; 11:5679.

14. Kingston B, Cutts RJ, Bye H, Beaney M, Walsh-Crestani G, Hrebien S, et al. Genomic profile of advanced breast cancer in circulating tumour DNA. Nat Commun. 2021; 12:2423.

16. Zhang JT, Liu SY, Gao W, Liu SM, Yan HH, Ji L, et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non-dmall vell lung cancer. Cancer Discov. 2022; 12:1690–701.
17. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018; 28:1747–56.

18. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018; 173:321–37.
19. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016; 17:31.

20. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21.
22. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017:PO.17.00011.

23. Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021; 595:432–7.

24. Fakhri N, Chad MA, Lahkim M, Houari A, Dehbi H, Belmouden A, et al. Risk factors for breast cancer in women: an update review. Med Oncol. 2022; 39:197.

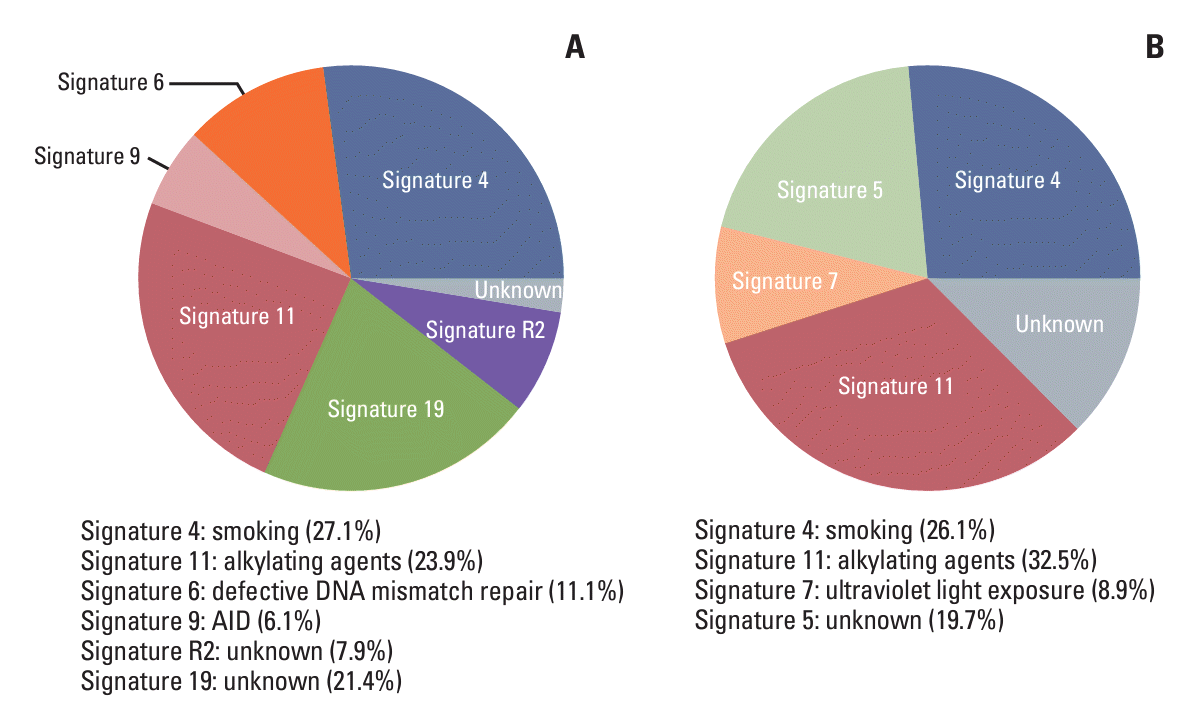
25. van Leeuwen FE, Ronckers CM. Anthracyclines and alkylating agents: new risk factors for breast cancer in childhood cancer survivors? J Clin Oncol. 2016; 34:891–4.

26. Olave MC, Graham RP. Mismatch repair deficiency: the what, how and why it is important. Genes Chromosomes Cancer. 2022; 61:314–21.

27. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017; 2017:PO.17.00073.

28. Sajjadi E, Venetis K, Piciotti R, Invernizzi M, Guerini-Rocco E, Haricharan S, et al. Mismatch repair-deficient hormone receptor-positive breast cancers: biology and pathological characterization. Cancer Cell Int. 2021; 21:266.

29. Cakan E, Gunaydin G. Activation induced cytidine deaminase: an old friend with new faces. Front Immunol. 2022; 13:965312.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download