Abstract
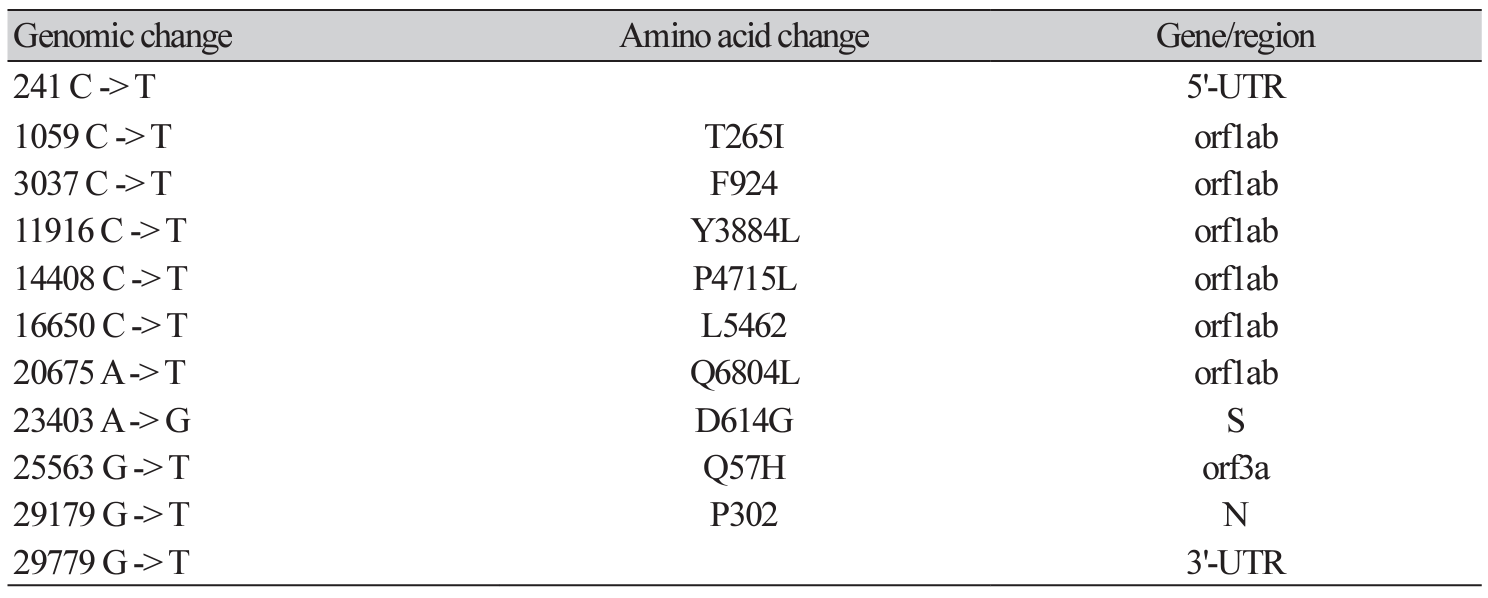
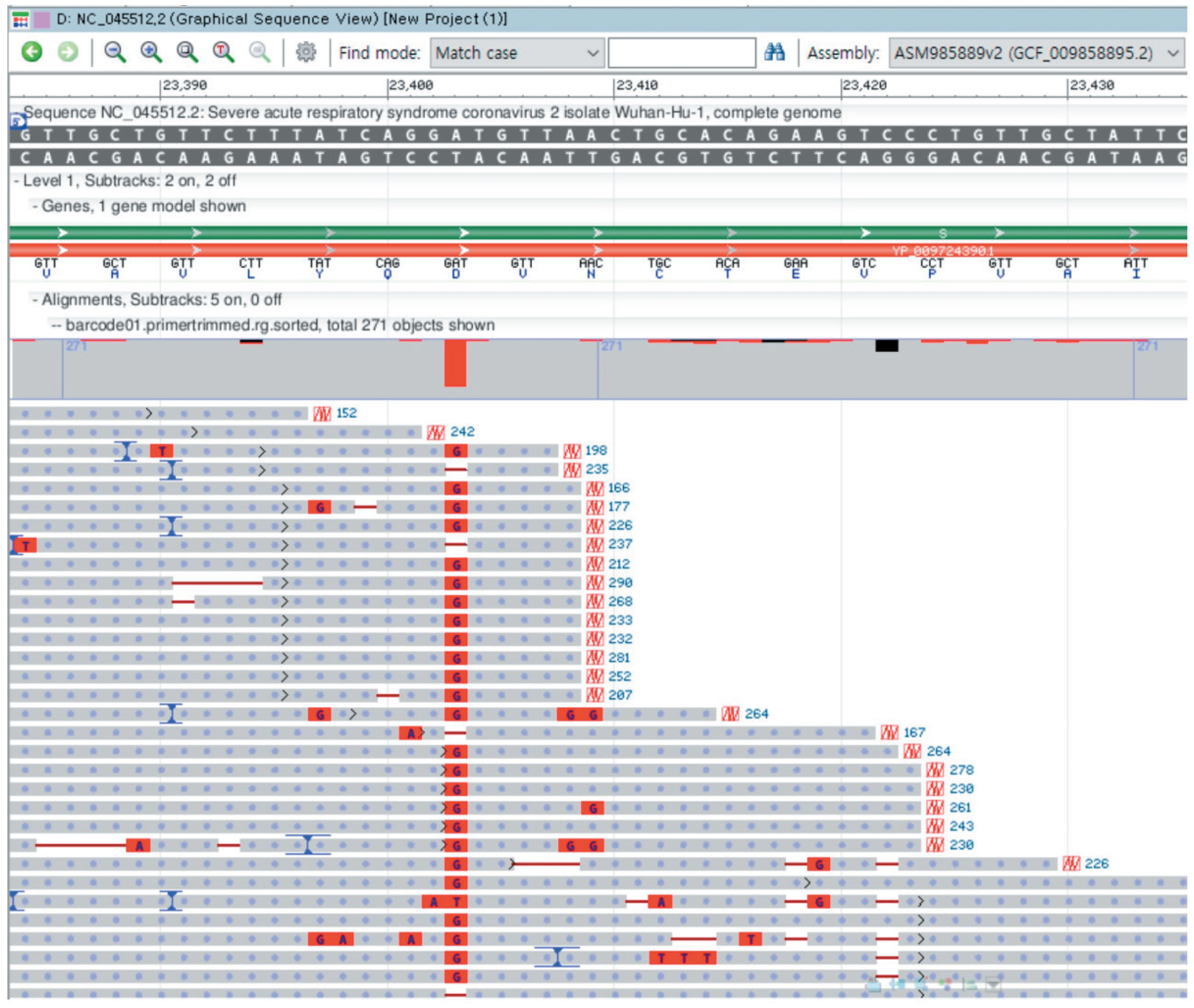
The application of whole genome sequencing on SARS-CoV-2 viral genome is essential for our understanding of the molecular epidemiology and spread of viruses in the community. The portable whole genome sequencer MinION (Oxford Nanopore Technologies, ONT, UK) could be feasibly used in a clinical microbiology laboratory without the need of vast resources or stringent operating conditions. We used the MinION sequencer to analyze the viral genome sequence of one SARS-CoV-2 strain. In June 2020, nasopharyngeal specimen from one patient was subjected to whole-genome analysis using the nCoV-2019 sequencing protocol v2 of ARTIC using the MinION sequencer. The ONT MinKNOW software, RAMPART tool, and Genome Workbench were used. We identified 11 nucleotide variants using the Wuhan-Hu-1 isolate (NC_045512.2) as the reference sequence. There were six nucleotide variants (T265I, F924, Y3884L, P4715L, L5462, and Q6804L) in the ORF1ab region, one variant (D614G) in the S gene, one variant (Q57H) in ORF3a, one variant (P302) in the N gene, and two variants in each the 5′-UTR and 3′-UTR. In this prolonged coronavirus disease 2019 (COVID-19) pandemic season, the MinION system that operates an amplicon-based whole-genome sequencing protocol could be a rapid and reliable sequencer without the need of cumbersome viral cultivation.
FUNDING
이 연구는 한림의대 산학협력단의 연구비 지원과 보건복지부의 재원으로 한국보건산업진흥원의 보건의료기술 연구개발사업 지원에 의하여 이루어졌습니다(Grant No. HI20C0071).
REFERENCES
1. Uhteg K, Carroll KC, Mostafa HH. Coronavirus detection in the clinical microbiology laboratory: are we ready for identifying and diagnosing a novel virus? Clin Lab Med 2020;40:459-72.
2. Burki T. Understanding variants of SARS-CoV-2. Lancet 2021;397:462.
3. Kim HM, Jeon S, Chung O, Jun JH, Kim HS, Blazyte A, et al. Comparative analysis of 7 short-read sequencing platforms using the Korean Reference Genome: MGI and Illumina sequencing benchmark for whole-genome sequencing. Gigascience 2021;10: giab014.
4. Kim JM, Park SY, Lee D, Kim JS, Park Y, Gwack J, et al. Genomic investigation of the coronavirus disease-2019 outbreak in the Republic of Korea. Sci Rep 2021;11:1-10.
5. Hourdel V, Kwasiborski A, Baliere C, Matheus S, Batejat CF, Manuguerra JC, et al. Rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100(TM) system. Front Microbiol 2020;11:571328.
6. Park AK, Kim IH, Kim J, Kim JM, Kim HM, Lee CY, et al. Genomic surveillance of SARSCoV-2: distribution of clades in the Republic of Korea in 2020. Osong Public Health Res Perspect 2021;12:37-43.
7. Bull RA, Adikari TN, Ferguson JM, Hammond JM, Stevanovski I, Beukers AG, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun 2020;11:6272.
8. Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. https:// doi.org/10.1101/2020.09.04.283077 BioRxiv 283077 [Preprint]. 2020 (last visited on 30 November 2021].
9. Tali SHS, LeBlanc JJ, Sadiq Z, Oyewunmi OD, Camargo C, Nikpour B, et al. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev 2021;34:e00228-20.
10. Park S, Kim DH, Lee WM, Ha JS, Jeon DS, Lee JH, et al. Experience at department of laboratory medicine during the COVID-19 outbreak in Daegu. Ann Clin Microb 2020;23:22531.
11. Hong KH, In JW, Lee J, Kim SY, Lee KA, Kim S, et al. Prevalence of a single-nucleotide variant of SARS-CoV-2 in Korea and its impact on the diagnostic sensitivity of the Xpert Xpress SARS-CoV-2 assay. Ann Lab Med 2022;42:96-9.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download