Abstract
Background: Blood volume is the most important parameter for an optimal blood culture; however, the effect of blood volume on blood culture is not clearly understood from patients with sepsis. Methods: Blood cultures were obtained from 1,049 patients (≥ 15 years old) who visited the emergency department (ED). Two sets of 20 mL each was collected from each patient, 12 mL of which was transferred to 2 and 10 mL FA Plus (aerobic) bottles (bioMérieux, USA) and the remaining into an FN Plus (anaerobic) bottle. Medical records were reviewed to confirm the diagnosis and clinical significance of the blood culture isolates. The positive rate and time-to- detection (TTD) were compared between the 2 and 10 mL groups. Results: Among the 2,098 sets collected, 612 sets (29.2%) were excluded due to inadequate (either too much or too little) blood volume. The positive rate of clinically significant pathogens was lower in the 2 mL group (6.1%) than in the 10 mL group (7.5%) (P = 0.003) among the 1,486 sets. However, there was no significant difference in the positive rate (11.0% vs. 12.5%, P = 0.152) and TTD (15.7 hours vs. 14.2 hours, P = 0.299) among the 585 (39.4%) patients with sepsis. Conclusion: The positive rate and TTD were similar between the 2 and 10 mL groups from patients with sepsis who visited the ED, suggesting a high concentration of bacteremia in this group. Therefore, a smaller blood volume should be carefully considered in patients with sepsis in the ED.
Blood amount is the most important parameter for optimal blood culture. About 20-30 mL should be collected for each set and be divided into aerobic and anaerobic bottles evenly [1,2]. The main reason to collect such a large volume of blood is based on the most important theory that bacteremia level is very low, sometimes less than < 1 CFU/mL [2,3]. However, we found out there are several pitfalls in this theory. First, the data supporting this dogma are too old, more than 30 years. There were no currently available automated continuous monitoring of blood culture systems (CMBCS) at that time. It is possible to collect 20-30 mL when they used the manual method, whereas 20 mL would be the maximum volume while using the current CMBCS. There are only a few studies regarding the effect of blood volumes in the era of CMBCS [4,5]. Second, there is a question whether the study design or statistical method was adequate for the previous studies [4,6-8]. We found the flaws that yield was exaggerated for the studies comparing 2 vs. 5 mL [6], or 5 vs. 10 mL [8]. When both-positive data is added, the difference between two groups becomes rather similar in both studies. Third, most of the previous studies did not focus on the sepsis patients. They just analyzed the positive cases of blood cultures in the laboratory, not by clinical diagnosis. If we include only the clinically diagnosed sepsis group, the result might be different.
The most serious limitation of blood cultures is the very low detection rate of pathogens even though we collect a large amount of blood volume. Second, the treatment modality is the same in many cases, regardless of the pathogens isolated [9,10]. Third, emotional stress is very high both for the phlebotomists and patients due to such a large blood volume sampling. Practically, it is very difficult to collect a large amount of blood in the emergency situation.
With these problems of the previous studies and poor clinical utility of blood cultures, we evaluated whether a smaller blood volume (2 mL) may replace the currently recommended blood volume (10 mL) for the sepsis patients group. The rationale to adopt 2 mL was based on the assumption that bacteremia level might be higher in the sepsis group than the other groups.
1. Patients
The patients (≥ 15 years old) who visited the emergency department (ED) of Gyeongsang National University Hospital during January to June, 2015 and were requested for blood cultures were enrolled for this study. This study was approved by the Institutional Review Board (IRB No. GNUH 2014-12-015), and written consent was exempted.
2. Blood cultures
After disinfection of skin with 0.5% chlorhexidine-alcohol, medical technicians drew 20 mL using a syringe for each set and put 2 and 10 mL into FA Plus (aerobic) bottles (bioMérieux, Durham, NC, USA) separately and the remaining into FN Plus (anaerobic) bottle. Two sets were collected for each episode in one patient. Blood culture bottles were incubated for 5 days at BacT/Alert 3D (bioMérieux) or until there was a positive signal. Bacterial identification was carried out using Vitek-2 system (bioMérieux, Marcy l'Etoile, France). Medical technicians were educated before or during this study for the proper skin disinfection and distribution of blood into each bottles.
3. Underlying infections and clinical significance of the isolates
Sepsis patients were screened and their underlying infections were registered by the database of electronic medical record (EMR) system retrospectively for the enrolled patients. Sepsis was defined bythe previous guideline [11]. All-cause-mortality in hospital was compared between blood culture-positive and -negative groups.
Streptococcus pneumoniae, β-hemolytic streptococci, Enterobacteriaceae, Neisseria meningitidis, Haemophilus influenzae, Pseudomonas aeruginsa, Acinetobacter baumannii, Staphylococcus aureus, Enterococcus species, and Salmonella species were primarily regarded as true pathogens. Staphylococcus epidermidis, other coagulase-negative staphylococci, α-hemolytic streptococci, non-hemolytic streptococci, diphtheroids, Bacillus species, and Micrococcus species were generally regarded as skin contaminants [1]. Final decision was made by an ED physician based on clinical manifestations and diagnosis in the EMR whether the isolates were true pathogens or skin contaminants. When the patient’s condition was improved by using the antibiotics effective for the isolates, or if the same pathogens were isolated in the other body site cultures, or if diagnosis was compatible with the isolates, then these isolates were considered as clinically significant pathogens.
4. Data analysis
All aerobic bottles were measured for weight. Then, we subtracted the weight of original bottle from the weight of the bottle after blood culture and divided it by the density of blood (1.055 g/mL) to calculate blood volume. Less than 1 mL or > 3 mL in the 2 mL group; < 8 mL or > 12 mL in the 10 mL group were regarded as inappropriate volume and excluded for the data analysis, because these cases would have significantly affected to the results [2].
Four different parameters were compared between 2 and 10 mL groups: first, the proportion of sepsis among the blood culture-requested patients; second, concomitant infections of these sepsis patients; third, the isolation rate of clinically significant pathogens or contaminants; fourth, time to detection (TTD). TTD is defined as the time interval between the entry of bottles into BacT/Alert 3D to the positive signal.
5. Statistical analysis
All data are presented as mean and standard deviation (SD). The independent samples t-test was used to test the difference of means by the independent two groups. Chi-square test was applied for calculating the difference of proportions by groups. McNemar test was carried out to observe the difference of positive rate and contamination rate between 2 and 10 mL groups. The paired t-test was used to analyze the difference of TTD. Statistical significance was defined as P < 0.050. IBM SPSS Statistics version 24.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis.
1. Blood volume
A total of 1,049 patients were enrolled for this study, which included 2,098 sets of blood cultures. After 612 sets were excluded due to the criteria of over- or under volume, 1,486 sets were included for analysis. Mean age was 64.1 ± 17.5 years, and males were 56.1%. The average blood volume revealed 2.3 ± 0.4 mL and 9.8 ± 0.8 mL in each group, respectively.
2. Positive rate
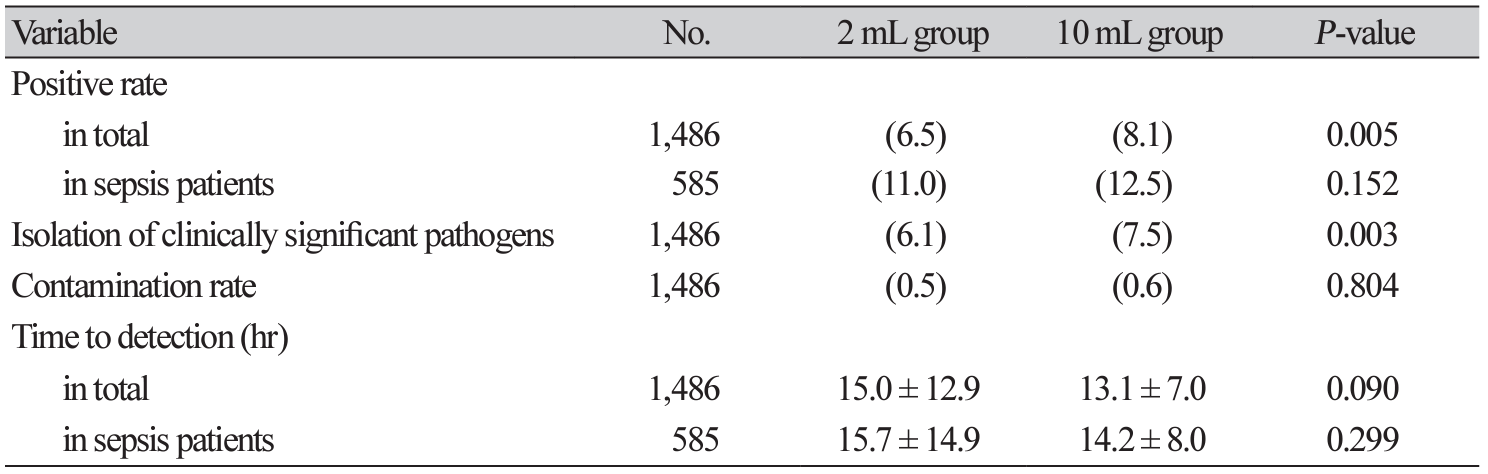
Microorganisms grew in 11.6%. The mortality was significantly higher in the ‘growth’ group compared with the ‘no growth’ group (12.8% vs. 7.7%, P = 0.022). Microorganisms were less frequently isolated in the 2 mL group compared with the 10 mL group (6.5% vs. 8.1%, P = 0.005) (Table 1). There was no significant difference in the contamination rate (0.5% vs. 0.6%, P = 0.804). Clinically significant pathogens were less commonly isolated in the 2 mL group compared with the 10 mL group (6.1% vs. 7.5%, P = 0.003).
3. TTD
There was no significant difference in TTD between the 2 mL group and 10 mL group (15.0 ± 12.9 hours vs. 13,1 ± 7.0 hours, P = 0.090) (Table 1). Among the sepsis group, there was also no significant difference of TTD (15.7 ± 14.9 hours vs. 14.2 ± 8.0 hours, P = 0.299).
4. Pathogens in the sepsis
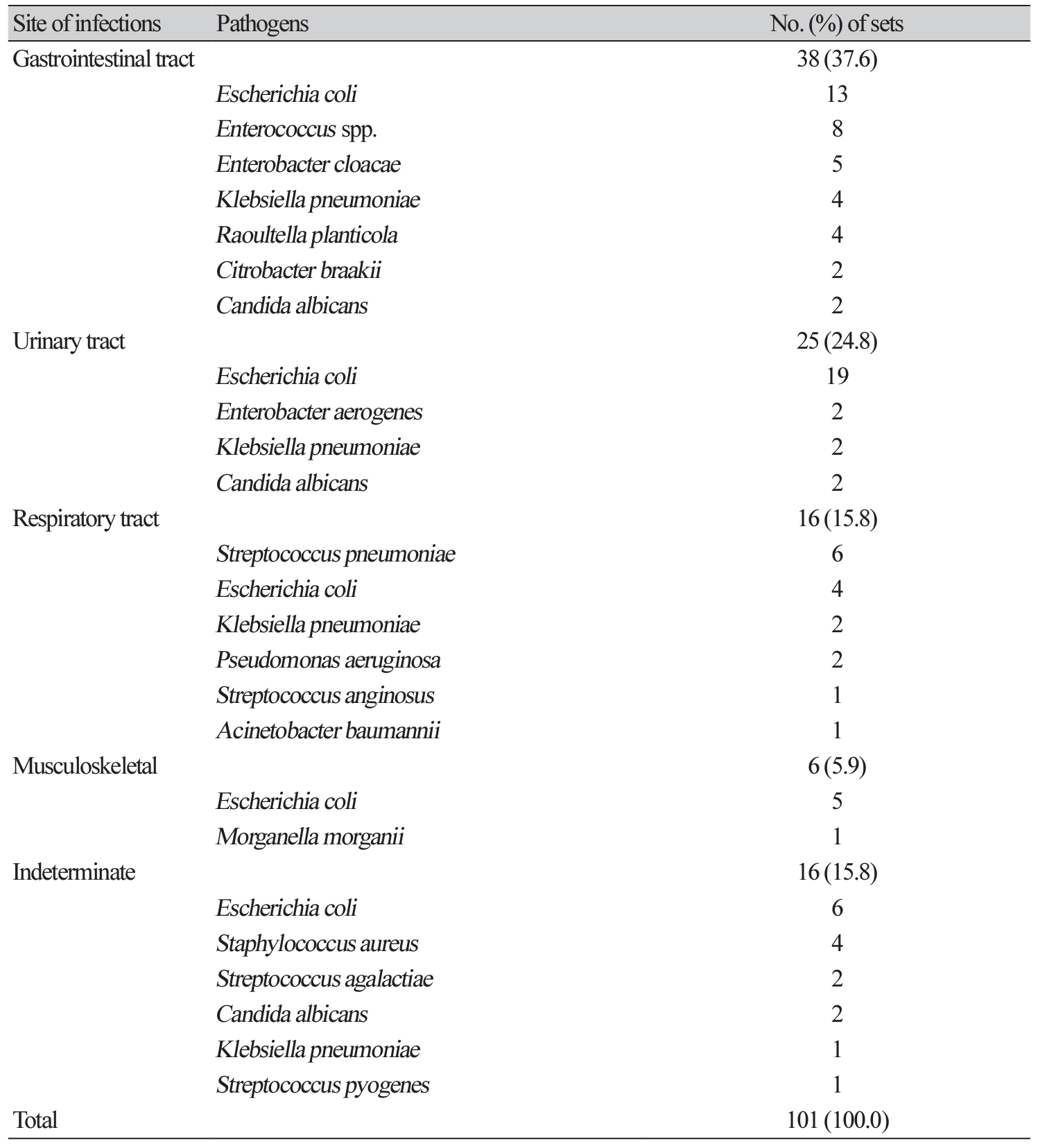
A total of 585 sets (39.4%) were taken from sepsis patients. Interestingly, there was no statistically significant difference of positive rate in this sepsis patients between 2 mL group and 10 mL group (11.0% vs. 12.5%, P = 0.152) (Table 1). A total of 101 sets grew pathogens among sepsis patients. Primary sites of infections were most common in gastrointestinal tract (37.6%), followed by urinary tract (24.8%), respiratory tract (15.8%), and musculoskeletal system (5.9%). Primary focus was indeterminate in 15.8% (Table 2). Escherichia coli (41.8%) was the most commonly isolated pathogen, followed by Klebsiella pneumoniae (8.9%), Enterococcus species (7.9%), and Streptococcus pneumoniae (5.9%) causing sepsis.
Although 20-30 mL of blood volume is recommended for each set [1,7], this amount is not easy to obtain, especially when the venous condition is not good. Taking the recommended blood volume is sometimes ignored in the very busy ED environment. The patients sometimes do not understand or cooperate to draw such a large amount of blood volume causing a serious argument. If we have a not-inferior yield with a small amount of blood volume (such as 2 mL), then we may expect many changes for the diagnosis of sepsis in the ED. This will relieve the tension between the patients and phlebotomists as well as enable faster fluid infusion and antibiotic administration, which is essential to save sepsis patient’s life [12]. Although there was a significant difference of positive rate between 2 mL group and 10 mL group in total, there was no difference specifically in the sepsis group.
Several studies indicated an increasing skin contamination rate with the increased blood volume, which caused unnecessary laboratory tests or longer admission days [2,7,13]. However, there was no difference in skin contamination rate between the two groups in this study. TTD was also similar regardless of blood volume, suggesting higher bacteremia level in the sepsis group. However, clinically significant pathogens were less commonly isolated in 2 mL group. Therefore, we should be careful to challenge the dogma: the more blood volume, the higher yield.
Gastrointestinal infections were the most common underlying infections among sepsis patients, and this finding is different from the previous studies [11]. E. coli comprised of almost half of pathogens in the sepsis patients, and the prevalence is much higher than the previous studies [4,11,14,15]. These findings indicate that there is a large gap for underlying infections and frequency of pathogens according to the study population and severity of sepsis patients.
There are several limitations in this study. First, the sample size (585 sepsis patients) is not enough to draw a strong conclusion. Second, this is a single-center, retrospective study. A multi-center prospective study may demonstrate more comprehensive data containing more sepsis cases. Third, the data of anaerobic bottle is not included due to difficulty of obtaining a large amount of blood. Fourth, we used the old definition of sepsis. As Sepsis-3 guideline was released in 2016 [16], it is necessary to analyze the data according to this new guideline. We observed a significant difference of positive rate as well as isolation of significant pathogens between two groups (2 vs. 10 mL) in total; therefore, the dogma (the more blood volume, the higher positive rate) seems still correct.
In conclusion, the smaller amount of blood volume (2 mL in this study) demonstrated a not-inferior microbiological outcome (positive rate and TTD) from the patients with sepsis. Therefore, smaller blood volume could be carefully considered for strongly suspected sepsis group in emergency situation.
REFERENCES
1. Clinical and Laboratory Standards Institute (CLSI). Principles and procedures for blood cultures; approved guideline. CLSI document M47-A. Wayne;PA, 2007.
2. Lamy B, Dargere S, Arendrup MC, Parienti JJ, Tattevin P. How to optimize the use of blood cultures for the diagnosis of bloodstream infections? A state-of-the art. Front Microbiol 2016;7:697.
3. Ilstrup DM and Washington JA, Jr. The importance of volume of blood cultured in the detection of bacteremia and fungemia. Diagn Microbiol Infect Dis 1983;1:107-10.
4. Cockerill FR 3rd, Wilson JW, Vetter EA, Goodman KM, Torgerson CA, Harmsen WS, et al. Optimal testing parameters for blood cultures. Clin Infect Dis 2004;38:1724-30.
5. Kim SC, Kim S, Lee DH, Choi SR, Kim JS. Effect of blood volume in standard anaerobic blood culture bottles of the BacT/ALERT 3D system used for the detection of pathogens and time to detection. PLoS One 2015;10:e0116728.
6. Tenney JH, Reller LB, Mirrett S, Wang WL, Weinstein MP. Controlled evaluation of the volume of blood cultured in detection of bacteremia and fungemia. J Clin Microbiol 1982;15:558-61.
7. Li J, Plorde JJ, Carlson LG. Effects of volume and periodicity on blood cultures. J Clin Microbiol 1994;32:2829-31.
8. Weinstein MP, Mirrett S, Wilson ML, Reimer LG, Reller LB. Controlled evaluation of 5 versus 10 milliliters of blood cultured in aerobic BacT/Alert blood culture bottles. J Clin Microbiol 1994;32:2103-6.
9. Shapiro NI, Wolfe RE, Wright SB, Moore R, Bates DW. Who needs a blood culture? A prospectively derived and validated prediction rule. J Emerg Med 2008;35:255-64.
10. Abe T, Tokuda Y, Ishimatsu S, Birrer RB. Usefulness of initial blood cultures in patients admitted with pneumonia from an emergency department in Japan. J Infect Chemother 2009;15:180-6.
11. Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence 2014;5:4-11.
12. Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Intensive Care Med 2013;39:165-228.
13. Self WH, Speroff T, Grijalva CG, McNaughton CD, Ashburn J, Liu D, et al. Reducing blood culture contamination in the emergency department: an interrupted time series quality improvement study. Acad Emerg Med 2013;20:89-97.
14. Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med 2003;348:1546-54.
15. Shin JH, Song SA, Kim MN, Lee NY, Kim EC, Kim S, et al. Comprehensive analysis of blood culture performed at nine university hospitals in Korea. Korean J Lab Med 2011;31:101-6.
16. Whittle J and Walker D. The new international sepsis guidelines (sepsis-3): the central message remains. Br J Hosp Med 2016;77:208-11.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download