Abstract
Background:We evaluated the performance of ASTA MicroIDSys (ASTA, Korea) and Bruker Biotyper (Bruker Daltonics, Germany) systems in the identification of bacterial isolates from clinical microbiology laboratory specimens during the study period. In addition, species for which the identification accuracy using MALDI-TOF MS systems was previously reported to be poor were also identified by comparing the MS results with those obtained using molecular identification. Methods:A total of 889 non-duplicated clinical isolates were included in this study. The results of ASTA MicroIDSys were compared with those of Bruker Biotyper; 16S rRNA sequencing was performed for the species for which results obtained using the two systems did not match. The sequences of rpoB , hisA, and/or recA for the clinical isolates of Acinetobacter species, Klebsiella species, and Burkholderia cepacia complex were analyzed and used as reference identifications. Results: The concordance rates for bacterial identification using ASTA MicroIDSys and Bruker Biotyper were 100% at the genus level and 98.3% at the species level for isolates belonging to the order Enterobacterales. Similarly, the concordance rates at the genus and species levels were 98.8% and 91.0% for glucose non-fermenting bacilli, 100% and 100% for gram-positive cocci, and 98.9% and 98.9% for other isolates, respectively. ASTA MicroIDSys was expected to correctly identify 97.9% of the 108,251 isolates identified in our clinical microbiology laboratory over the past 5 years. Conclusion:ASTA MicroIDSys showed excellent performance in bacterial identification for most of the clinically relevant species. Further extension of the database could improve the identification accuracy of ASTA MicroIDSys.
REFERENCES
1. Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med 2003;115:529-35.
2. Kootallur BN, Thangavelu CP, Mani M. Bacterial identification in the diagnostic laboratory: how much is enough? Indian J Med Microbiol 2011;29:336-40.
3. Park Y, Lee Y, Kim M, Choi JY, Yong D, Jeong SH, et al. Recent trends of anaerobic bacteria isolated from clinical specimens and clinical characteristics of anaerobic bacteremia. Infect Chemother 2009;41:216-23.
4. Brook I. The role of anaerobic bacteria in bacteremia. Anaerobe 2010;16:183-9.
5. Couzinet S, Jay C, Barras C, Vachon R, Vernet G, Ninet B, et al. High-density DNA probe arrays for identification of staphylococci to the species level. J Microbiol Methods 2005;61:201-8.
6. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761-4.
7. Clinical and Laboratory Standards Institute (CLSI). Interpretative criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. MM18-A. Wayne PA: 2007.
8. Sauer S, Kliem M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol 2010;8:74-82.
9. Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009;49:543-51.
10. Carbonnelle E, Mesquita C, Bille E, Day N, Dauphin B, Beretti JL, et al. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin Biochem 2011;44:104-9.
11. Drancourt M. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin Microbiol Infect 2010;16:1620-5.
12. Pavlovic M, Konrad R, Iwobi AN, Sing A, Busch U, Huber I. A dual approach employing MALDI-TOF MS and real-time PCR for fast species identification within the Enterobacter cloacae complex. FEMS Microbiol Lett 2012;328:46-53.
13. Sousa C, Botelho J, Grosso F, Silva L, Lopes J, Peixe L. Unsuitability of MALDI-TOF MS to discriminate Acinetobacter baumannii clones under routine experimental conditions. Front Microbiol 2015;6:481.
14. Fehlberg LC, Andrade LH, Assis DM, Pereira RH, Gales AC, Marques EA. Performance of MALDI-ToF MS for species identification of Burkholderia cepacia complex clinical isolates. Diagn Microbiol Infect Dis 2013;77:126-8.
15. Angeletti S. Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J Microbiol Methods 2017;138:20-9.
16. Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnosticsidentification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol 2012;93:965-74.
17. Lee Y, Sung JY, Kim H, Yong D, Lee K. Comparison of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform, ASTA MicroIDSys, with Bruker Biotyper for species identification. Ann Lab Med 2017;37:531-5.
18. Lee H, Park JH, Oh J, Cho S, Koo J, Park IC, et al. Evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for the identification of yeast isolation. J Clin Lab Anal 2019;33:e22685.
19. Kim D, Ji S, Kim JR, Kim M, Byun JH, Yum JH, et al. Performance evaluation of a new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, ASTA MicroIDSys system, in bacterial identification against clinical isolates of anaerobic bacteria. Anaerobe 2020;61:102131.
20. Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 2009;155:2333-41.
21. Papaleo MC, Perrin E, Maida I, Fondi M, Fani R, Vandamme P. Identification of species of the Burkholderia cepacia complex by sequence analysis of the hisA gene. J Med Microbiol 2010;59:1163-70.
22. Payne GW, Vandamme P, Morgan SH, Lipuma JJ, Coenye T, Weightman AJ, et al. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol 2005;71:3917-27.
23. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 2012;62:716-21.
24. Ling TK, Liu ZK, Cheng AF. Evaluation of the VITEK 2 system for rapid direct identification and susceptibility testing of gram-negative bacilli from positive blood cultures. J Clin Microbiol 2003;41:4705-7.
25. Mera RM, Miller LA, Amrine-Madsen H, Sahm DF. Acinetobacter baumannii 2002-2008: increase of carbapenem-associated multiclass resistance in the United States. Microb Drug Resist 2010;16:209-15.
26. Shim BS, Kim CS, Kim ME, Lee SJ, Lee DS, Choe HS, et al. Antimicrobial resistance in community-acquired urinary tract infections: results from the Korean antimicrobial resistance monitoring system. J Infect Chemother 2011;17:440-6.
27. Jeong S, Hong JS, Kim JO, Kim KH, Lee W, Bae IK, et al. Identification of Acinetobacter species using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Ann Lab Med 2016;36:325-34.
28. Speert DP. Advances in Burkholderia cepacia complex. Paediatr Respir Rev 2002;3:230-5.
29. Marioni G, Rinaldi R, Ottaviano G, Marchese-Ragona R, Savastano M, Staffieri A. Cervical necrotizing fasciitis: a novel clinical presentation of Burkholderia cepacia infection. J Infect 2006;53:e219-22.
30. Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, LiPuma JJ, et al. Burkholderia cenocepacia sp. nov.-a new twist to an old story. Res Microbiol 2003;154:91-6.
31. Ragupathi NKD and Veeraraghavan B. Accurate identification and epidemiological characterization of Burkholderia cepacia complex: an update. Ann Clin Microbiol Antimicrob 2019;18:1-10.
32. Lacroix C, Gicquel A, Sendid B, Meyer J, Accoceberry I, Francois N, et al. Evaluation of two matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for the identification of Candida species. Clin Microbiol Infect 2014;20:153-8.
33. Girard V, Mailler S, Polsinelli S, Jacob D, Saccomani MC, Celliere B, et al. Routine identification of Nocardia species by MALDI-TOF mass spectrometry. Diagn Microbiol Infect Dis 2017;87:7-10.
Table 1
Comparison of bacterial identification between ASTA MicroIDSys and Bruker Biotyper for 418 Enterobacterales isolates
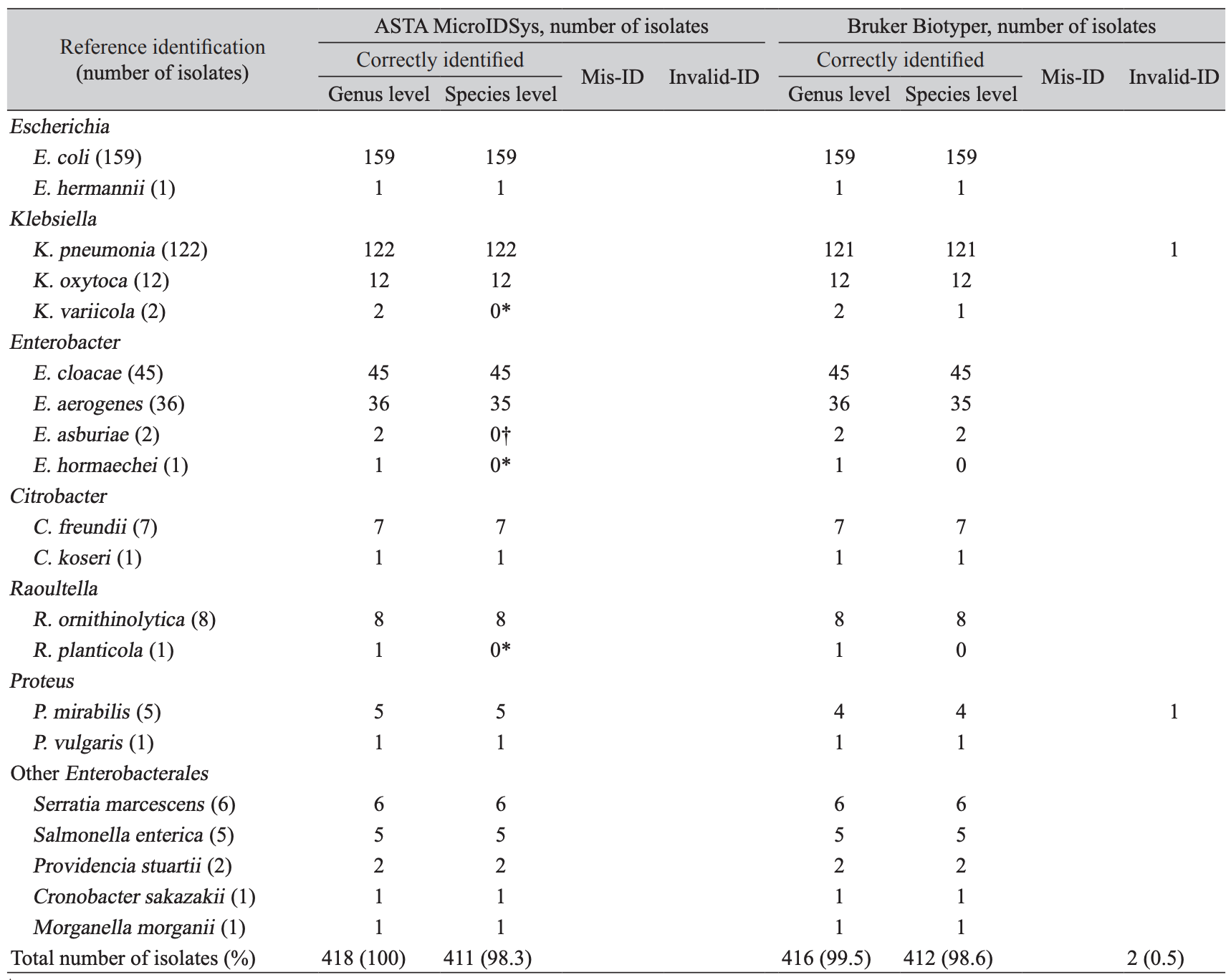
Table 2
Comparison of bacterial identification between ASTA MicroIDSys and Bruker Biotyper for 167 glucose non-fermenting bacilli isolates

Table 3
Comparison of bacterial identification between ASTA MicroIDSys and Bruker Biotyper for 216 Gram-positive cocci isolates

Table 4
Comparison of bacterial identification between ASTA MicroIDSys and Bruker Biotyper for 88 other isolates





 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download