Abstract
Musculoskeletal infections (MSKI) present a significant health challenge, with a rising incidence linked to the aging population and advancements in orthopedic surgical care. Staphylococcus aureus is the most prevalent pathogen associated with orthopedic infections. The conventional culture method for identification of pathogen frequently lacks accuracy and is challenged by false-positive or false-negative results. Inflammatory markers such as the erythrocyte sedimentation rate and C-reactive protein are not site-specific or accurate, as they can be confounded by other medical conditions. Identifying the dominant pathogen and monitoring treatment response following surgical debridement and antibiotics therapy continues to pose challenges. Understanding the pathogenesis of MSKI is crucial for the development of innovative diagnostics and alternative therapeutics. S. aureus immune evasion stands out as a key component of the pathogenic mechanism, complicating clinical decisions. Other unique mechanisms such as biofilm and abscess formation, as well as osteocyte-lacuno canalicular network invasion, underscore the need for aggressive debridement and the complete removal of infected implants and bone tissues. Ongoing efforts focus on exploring and developing innovative diagnostics, such as serum immunoassays, next-generation sequencing of infected tissue, transcriptomics of peripheral blood mononuclear cells, and serum proteomics. These endeavors offer promising avenues for improved diagnostics, medical management, and innovative therapeutics for MSKI.
Deep musculoskeletal infections (MSKI) are major orthopedic challenges, which have steadily increased [1-4]. This includes infection of the bone (osteomyelitis), native joints (septic arthritis), soft tissue (cellulitis, abscess, necrotizing fasciitis), bursa and tendon (infected bursitis and tendonitis), and implant-associated periprosthetic infections (Fig. 1). MSKI can arise from three main causes: adjacent spread of infection from surrounding tissue, direct bone trauma from injury, or hematogenous spread [4]. The increasing incidence of recurrent MSKI is a major healthcare burden worldwide, leading to higher rates of unplanned readmissions and reoperations [5,6]. Thus, there is a tremendous need for developing an improved diagnostic to identify patients with MSKI, for whom more prompt and aggressive interventions are warranted [7-9]. Treatment for MSKI typically involves surgically removing the infected source, such as an infected implant, bone, or soft tissues, followed by a course of intravenous and/or oral antibiotics [10-12]. MSKI can be challenging to differentiate from other inflammatory conditions, such as acute exacerbation of degenerative or post-traumatic arthritis, inflammatory arthritis (e.g., rheumatoid arthritis), crystal arthropathies (e.g., gout), and neuroarthropathy [13,14]. Many clinicians utilize nonspecific serum inflammatory markers, such as the C-reactive protein and erythrocyte sedimentation rate, which are not reliable nor specific for infection [7-9]. Conventional culture to identify main pathogen has been challenged by false-positive or false-negative cultures, especially in the setting of chronic antibiotics use and recurrent polymicrobial infection [3,4,7-9]. Among various pathogens affecting the musculoskeletal system, Staphylococcus aureus is the most prevalent pathogen in various MSKI, such as infected diabetic foot ulcers (DFU; 46%–68%), prosthetic joint infection (PJI; >50%), fracture related infection (FRI; 42%–57%), spine infection (50%–65%), hand infection (60%–80%), and septic arthritis of the native joints (>50%) [5,6,8,15-19]. Other pathogens associated with MSKI include coagulase-negative Staphylococci (Staphylococcus epidermidis), Streptococcus, Enterococcus, and Gram-negative microbials [3,4]. Improved diagnostics for MSKI, especially for S. aureus infection, would provide tremendous clinical benefits for the management of acute and recurrent MSKI. This article focuses on addressing clinical challenges and advancements in diagnosing MSKI.
S. aureus is a commensal microorganism that ubiquitously resides on the skin, mucous membranes, and various surfaces in humans [4,7,19]. Most human infants have a colonization rate of over 50% during the first 2 months of life [20,21]. Among adults, around 20% are nasal carriers of S. aureus, and 30% carry it intermittently [20]. Though harmless for most healthy people, S. aureus has the potential to become pathogenic. This shift can lead to a wide range of infections, from minor skin infections to life-threatening conditions like endocarditis, pneumonia, osteomyelitis, and bacteremia [22,23]. It stands as the most predominant and most destructive pathogen in orthopedic infections. The transition from commensal to pathogen can occur under certain conditions, such as an immune-compromised state or a breach in the host’s biologic barrier due to injury or surgery [3,4]. Upon entering the host, S. aureus encounters a series of attacks from the host’s immunologic defense mechanisms, against which it has evolved counteractive or evasive responses [22,23]. In addition to immune evasion mechanisms, the invasion of the musculoskeletal system by S. aureus exhibits distinctive features, including (1) biofilm formation on bone and prosthetic implants, (2) abscess formation, and (3) colonization of the submicron osteocyte-lacuno canalicular network (OLCN) (Fig. 2) [3,4,24,25]. Especially a biofilm can serve as a protective shield or a biologic barrier, which effectively insulates S. aureus from the host’s immune surveillance and antibiotics [3,4,25]. Understanding the clinical factors and molecular mechanisms that govern the invasion and survival strategies of S. aureus will enhance our understanding of the pathogenesis and suggest improved diagnostics for S. aureus-associated orthopedic infections.
The common immunogenic S. aureus antigens associated with MSKI can be categorized as follows: (1) iron-regulated surface determinant protein (IsdA, IsdB, IsdH); (2) cell wall enzyme bifunctional autolysin (amidase [Amd], glucosaminidase [Gmd]); (3) secretory toxins (α-hemolysin, chemotaxis inhibitory protein of S. aureus [CHIPS], Staphylococcal complement inhibitor [SCIN]); and (4) adhesins (clumping factor [ClfA, ClfB], fibronectin-binding protein A [FnBPA]) [7,26]. Antibodies against S. aureus antigens are generated or boosted during an active infection and their concentration usually increase [7,9,20,26,27]. We employed a multi-antigen Luminex immunoassay to explore the humoral immune proteome of patients with S. aureus-associated MSKI. Based on our analysis of 110 MSKI patients, we noted significantly increased anti-S. aureus serum antibodies against IsdB, IsdH, and SCIN in the human serum samples during the active phase of S. aureus-associated MSKI. In particular, elevated levels of anti-IsdB and anti-IsdH antibodies have been reported to be associated with more severe cases of S. aureus-associated MSKI [1,7,9].
Like numerous other microorganisms, the acquisition of essential minerals from the host is crucial for the survival of a pathogen. The iron acts as a cofactor for enzymes in essential metabolic processes [28,29]. S. aureus utilizes Isd system to scavenge iron from the host’s hemoglobin. The Isd pathway consists of multiple proteins that work in concert to acquire heme. Three receptor proteins, IsdA, IsdB, and IsdH, are anchored to the cell wall. Among them, S. aureus utilizes IsdB and IsdH to capture hemoproteins at its bacterial surface [28,30]. Hemoglobin is captured by IsdH and IsdB through a widely conserved near iron transport (NEAT) domains [28]. The IsdB extracts heme from hemoglobin and then transports it to downstream Isd proteins, IsdA and IsdC, through which heme is transported into the cell and degraded, thereby releasing iron for utilization by S. aureus (Fig. 3) [28-30]. IsdB has two NEAT domains: NEAT1 binds to hemoglobin, and NEAT2 binds and transports the heme. Particularly high titers of anti-IsdB have been reported to be associated with more severe S. aureus MSKI with high mortality and has been investigated as a potential vaccine candidate [31]. However, a phase 2/3 clinical trial for Merck’s IsdB-targeting vaccine (V710) was halted due to a concerning rise in patient mortality associated with multiple organ failure and sepsis in the vaccinated group [32]. Some hypothesized that vaccination may lead to the generation of non-neutralizing anti-IsdB antibodies, potentially impeding the action of neutralizing anti-IsdB antibodies and subsequently leading to the failure of clearance by our immune system. Eventually it disseminated to multiple organs, causing septic death [3,4,26,31,33]. The precise mechanism of failure continues to be a topic of ongoing debate. Some suggest that a more selective targeted vaccine against the IsdB heme-binding domain may enhance the success of protective immunization [31].
S. aureus has developed various strategies to effectively evade both innate and adaptive host defenses, which involve cell-mediated responses led by T-cells and humoral antibody responses mediated by B cells. While anti-S. aureus antibodies are commonly found in all humans due to prior exposure, their presence alone does not ensure protection for the host.
Ongoing research has revealed the complex interactions between various virulence factors of S. aureus and the host’s defense mechanisms, and it continues to reveal new insights [3,4,23,26]. S. aureus secretes various toxins and virulence factors which combat against host’s innate and adaptive immunity. The secretory toxin and virulence factors include adhesins, immunomodulatory proteins, toxins and superantigens [4,34]. S. aureus secrets pore-forming toxins, such as α-hemolysin, β-hemolysin, γ-hemolysin (HlgAB and HlgCB), leucocidin A/B (LukAB) and Panton-Valentine leukocidin that directly destroy neutrophil, macrophages and other antigen-presenting cells by damaging their cellular membranes [35,36]. Superantigens such as S. aureus enterotoxin B and C (SEB, SEC) and toxic shock syndrome toxin 1 (TSST1) induce a shift in M2 macrophage polarization, foster the formation of myeloid-derived suppressor cells, and disrupt antigen presentation and cytokine production, ultimately impacting the activation of T and B cells. S. aureus utilizes several cell-wall associated microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) to facilitate its internal invasion of macrophages and neutrophils (ClfA, ClfB, FnBPA, fibronectin-binding protein B [FnBPB]) [37,38]. S. aureus also interferes complement-mediated opsonization and phagocytosis through secretion of virulence proteins, such as CHIPS, SCIN, coagulase, and extracellular fibrinogen binding protein (Efb) (Fig. 4) [4,37,38].
Antibody productions against a broad spectrum of S. aureus antigens are very common in healthy individuals [4,20,26]. Unfortunately, the presence of pre-existing antibodies (“humoral immune proteome”) does not necessarily translate to immunity against S. aureus infection. Adaptive immunity encompasses T cell-mediated cellular responses and humoral antibody responses mediated by B cells. As mentioned earlier, superantigens like TSST1 can skew T cell activation, resulting in compromised memory T cell responses and induction of anergy [39]. S. aureus also secretes the protein SpA, which has the ability to binds to the Fc and Fab domains of specific antibodies. This interaction hinders antibody-mediated phagocytosis while simultaneously triggering apoptosis in proliferative B cells [40]. In addition, the S. aureus enzyme staphylokinase (Sak) has the capability to directly degrade IgG (Fig. 4) [41]. Some individuals may be more prone to S. aureus infections than others, potentially because of differences in the protective versus susceptible components of their immune proteome. Comprehending the functional dynamics between the protective and pathogenic aspects of an individual’s antibody response is crucial for the advancement of passive or active immunotherapies against S. aureus. Immunotherapies could potentially function as an adjuvant to antibiotics in the treatment of orthopedic infections. Despite numerous preclinical validations, none of the trials to produce immunization against S. aureus have succeeded yet.
In addition to the previously mentioned immune evasion mechanisms, S. aureus has evolved distinct pathogenic traits to survive in the host’s musculoskeletal system.
Formation of a biologic barrier on the bone or implant can effectively shield S. aureus against various immune responses and antibiotics. Biofilm formation occurs in four stages which include bacterial cell attachment, proliferation, biofilm maturation, and detachment [3]. S. aureus adheres to a substrate, such as implant material or necrotic bone, using surface adhesion molecules known as MSCRAMM. These proteins, such as FnBPA, FnBPB, SpA, ClfA, ClfB, situated on the microbial cell surface, exhibit the capability to bind to ligands found in the extracellular matrix, such as fibronectin, fibrinogen, and collagen [3,26]. The attached cells proliferate to amplify the bacterial population and commence the production of a matrix of extracellular polymeric substances (EPS) to shield the multi-layered community [3,26,42]. EPS is generally composed of polysaccharides, proteins, and nucleic acids, which mediate adhesion, provide mechanical stability, and retain essential nutrients and enzymes [3,26,42]. Subsequently, the biofilm detaches, allowing bacterial cells colonize new regions within the host. The acquisition of antimicrobial resistance gene within the biofilm is highly dynamic, primarily attributed to active horizontal gene transfers [3,4]. In addition to providing prolonged bacterial survival, biofilm can induce damage to the adjacent bone by triggering bone resorption through a combination of inflammatory and bacterial factors. It stimulates the release of inflammatory cytokines, including TNF-α, IL-1, and IL-6, resulting in the activation and differentiation of osteoclasts [43]. For orthopedic surgeons, the aggressive removal of biofilm from implanted hardware and surrounding necrotic tissue is crucial for eliminating and preventing recurrent infections. However, physical removal of biofilm from implant hardware by irrigation alone has faced limited success [44]. Therefore, complete hardware removal, coupled with thorough debridement, is considered the gold standard to minimize recurrent infection in implant-associated MSKI (Fig. 2).
S. aureus possesses the unique capability to establish chronically infect bone marrow and peri-implant soft tissue through the formation of Staphylococcal abscess communities (SAC) during MSKI [3,4,45]. An abscess is generally a host-induced mechanism for infection control, and it can develop as early as 4 days after the initiation of infection [45,46]. The SAC restrict blood flow to the core of infection and confers resistance to antibiotics and host immune responses against pathogens at the core [45,47]. This SAC within bone during osteomyelitis is also described as Brodie abscesses [47]. S. aureus first creates a protective shield around itself. This shield, which is a fibrous pseudocapsule, is formed through the activity of coagulase and von Willebrand factor-binding protein (vWbp). These proteins activate prothrombin, which then convert fibrinogen to fibrin, a key component of the capsule. Additionally, proteins ClfA and ClfB directly bind to fibrinogen, helping to promote the fibrin margin formation at the periphery of the abscess [46,48,49]. During S. aureus colony formation, a significant number of immune cells undergoes necrosis through a combination of direct and indirect killing. Over weeks, as the SAC matures, immune cells become trapped at the edges, unable to reach the bacteria at the center where they continue to multiply unchecked [46,48,49]. Therefore, the host’s immune system alone is unable to completely eradiate SACs and require surgical debridement to eliminate all abscesses (Fig. 2) [3,4,45].
We recently observed the colonization of the OLCN by S. aureus in a patient with chronic osteomyelitis of the infected diabetic foot (Figs. 2, 5) [24]. This phenomenon was first observed during a comprehensive examination of infected bone using transmission electron microscopy in an experimental murine model of chronic S. aureus osteomyelitis [50]. It revealed S. aureus invasion of canaliculi perpendicular to the medullary canal and subsequent colonization of lacunar spaces, devoid of osteocytes [50]. In contrast to bacteria in the bone marrow, which are surrounded by neutrophils, S. aureus inside the OLCN is encased by the dense mineral matrix of cortical bone, providing complete shielding from immune cells [50]. These observations indicate that S. aureus could thrive within the OLCN, taking advantage of abundant nutrient supply, and effectively evading immune responses [3,50]. The identification of OLCN invasion poses challenges for managing implant-associated osteomyelitis. It’s unclear whether current methods can achieve complete removal of infected OLCN, and if standard antibiotics therapy can effectively target S. aureus embedded within the bone cortex.
Novel diagnostic tests such as synovial fluid α-defensin enzyme-linked immunosorbent assay (ELISA), nucleic acid-based test, such as the next-generation sequencing (NGS) for serum, synovial fluid and tissue cultures have been proposed to improve sensitivity and accuracy in identifying microorganism [19]. However, clinical applications of those molecular diagnostics have been constrained by various factors such as access, cost, and mixed reports regarding their clinical efficacy. Given these challenges, alternative diagnostics continue to be developed.
Application of immunoassay for identifying S. aureus in cases of MSKI presenting with clinical symptoms and signs of orthopedic infection have been investigated. As serum immunoassay for S. aureus can be confounded by the pre-existing anti-S. aureus IgG resulting from prior exposure, the use of antibody-secreting cells (ASC), also known as plasmablasts, generated during active infections could offer a more reliable method for detecting active or acute S. aureus infection [1,2,7,9] As ASC are present in the blood only during active infections, they emerge early in a nascent infection and decline when the host response or therapeutic intervention has been successful [1,2,7,9]. By harvesting the ASC and placing them in culture media free of pre-existing antibodies, we were able to measure the newly synthesized antibody reflecting only the active or ongoing infection. We demonstrated that harvested ASCs secretes IgG in vitro, creating a novel analytic fluid, medium-enriched for newly synthesized antibody (MENSA). This innovative approach enables the antibody-based diagnosis of acute S. aureus infections [1,2,7,9]. Using a multiplex immunoassay (Luminex) that simultaneously measures immunoglobulin responses against 10 different S. aureus antigens, we noted high titers of antigen during active infection [7,9,27]. These titers resolved by 6 weeks with the administration of long-term antibiotics. A sustained decrease in antibody levels was observed during the resolution of the infection, while elevated titers were noted in cases of recurrent infection [7,9,27]. With the initial success, we expanded our study to include 110 MSKI patients that included DFU, PJI, FRI, septic arthritis of the native joints, spine and hand infections. We noted that anti-IsdB, IsdH, and SCIN antibodies showed a strong correlation with culture identified S. aureus MSKI in MENSA as well as in serum [9,27]. While the MENSA immunoassay showed a positive correlation with active MSKI, it did not exhibit a distinct improvement over the serum-based immunoassay [28-31]. While the serum-based immunoassay may encounter challenges from pre-existing antibodies against S. aureus, heightened pathogenic activity could potentially enhance adaptive immune responses enough to differentiate between increased immunity against S. aureus versus a dormant state [1]. The vital role of IsdB for S. aureus MSKI pathogenesis has been reported [28-31]. In particular, the anti-IsdB antibody has been recognized as a “pathogenic antibody” (vs. a protective antibody), exhibiting high titers in MSKI patients with adverse clinical outcomes [51]. Further investigation is warranted to explore the utility of anti-IsdB, IsdH and SCIN in diagnosing and monitoring treatment response for S. aureus specific MSKI.
The conventional diagnostic method of microbial culture has limitations, such as inadequate sampling and contamination associated with instruments or improper handling of specimen [3,7,52]. This can result in false-positive or false-negative growth, and ultimately may not provide a clinically useful information to guide antibiotics therapy [3,8]. Previous studies have shown 46.2% rate of recurrent infection within 3 months after debridement and antibiotics therapy of infected DFUs [7]. Given the challenges associated with accurate identification of dominant pathogens in polymicrobial infections with high recurrent infection rates, clinical application of molecular diagnostics has been investigated for MSKI. NGS is a high-throughput diagnostic method that enables the simultaneous sequencing of billions of DNA fragments in parallel. Two broad methodologic approaches to implement NGS are: (1) 16S amplicon targeted NGS and (2) shotgun metagenomics, which sequences all the genomic DNA from a given sample [52,53]. When using NGS to determine bacterial genome, the genome is first fragmented into multiple pieces, which are sequenced and assembled. The resulting genome assembly is subsequently compared to a reference database for organism identification. Clinical application of NGS in identifying pathogen associated with PJI after hip, knee, or shoulder arthroplasty has been explored [54-56]. We investigated 30 patients with infected DFU and comparatively analyzed NGS with standard culture [52]. We noted complete concordance of conventional culture and NGS in 14 cases (46.7%), partial concordance in eight cases (26.7%) and discordance in eight cases (26.7%). In the conventional culture, S. aureus (58.6%) and coagulase-negative Staphylococcus (24.1%) were the most identified bacterial species, followed by Corynebacterium striatum (17.2%) and Enterococcus faecalis (17.2%). In NGS, Finegoldia magna (44.8%), was the most identified organism followed by S. aureus (41.4%) and Anaerococcus vaginalis (24.1%). Overall, NGS detected a greater polymicrobial presence in each sample than standard culture [52]. However, whether the identified microorganisms presented the true pathogen, bystander (commensal), or contaminant was not clear. Other authors who utilized NGS for diagnosis of PJI reported conflicting results [53,55,56]. One advantage of NGS is its ability to identify antibiotic-resistance gene, which may be useful for guiding antibiotics choice. Since the identification of antibiotic-resistance gene is relatively fast, NGS allows earlier initiation of targeted antibiotics and provides a significant advantage over the current standard culture guided clinical care. Despite those advantages, the widespread application of NGS faces challenges in clearly differentiating the dominant pathogen from other commensals in polymicrobial infections, as well as high costs. Further investigation is warranted to explore and optimize its clinical utilization.
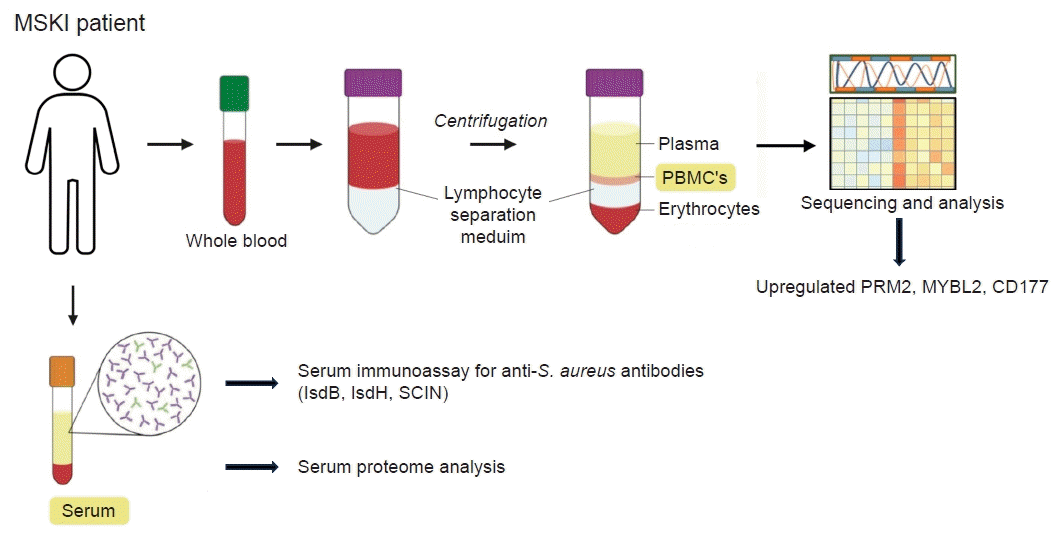
Obtaining tissue sample frequently requires invasive procedures such as aspirating native joint, image-guided sampling, or surgery. We have explored potential biomarkers from peripheral blood mononuclear cell (PBMC) and serum isolated from the blood samples of patients with MSKI. The evolving genetic transcriptions of PBMC in individual with MSKI can provide valuable insights for diagnosis and monitor treatment responses. We investigated transcriptome profile of 21 patients with S. aureus infected diabetic foot who underwent surgical debridement followed by 6 weeks course of antibiotics therapy. PBMC were isolated from the patients’ blood samples collected at 0 and 8 weeks after therapy, and their genetic transcriptomes were comparatively analyzed. An increased expression of IGHG1, IGHG2, IGHG3, IGLV3-21, IGLV6-57 genes was noted during active infection at 0 week compared to that of 8 weeks after therapy. Most notably the ribonucleotide reductase regulatory subunit M2 (RRM2), a transcription factor in the Myb-related protein B (MYBL2), and a neutrophil-specific cell surface glycoprotein (CD177) were upregulated at the initial phase of infection and downregulated at 8 weeks. RRM2 is a protein that is a subunit of the ribonucleotide reductase enzyme, which is involved in synthesis and damage repair of DNA. It has been suggested as a biomarker for certain cancer, such as hepatocellular carcinoma and lung adenocarcinoma [57,58]. Alongside RRM2, MYBL2 and the CD177 gene exhibited heightened expression levels during the active phase of infection (0 week) and returned to normal levels at 8 weeks post-therapy. We can utilize this information to validate PBMC or serum proteomics in patients, paving the way for the development of novel diagnostics for MSKI (Fig. 6). In addition, PBMC transcriptomes may also identify S. aureus specific infection. Previous studies have shown that the heavy chain variable region of IGHV1-69 gene commonly encodes human monoclonal antibodies targeting IsdB-NEAT2 binding site. One study demonstrated that IGHV1-69 encoded antibodies contribute to a protective immune response against S. aureus infection [28]. The upregulation of IGVH1-69 gene may serve as a potential biomarker in diagnosing S. aureus specific MSKI and warrants further investigation.
Despite recent advances in diagnostics and antibiotics therapy, the management of MSKI remains challenging, with a persistent increase in incidence. S. aureus stands out as the most common pathogen, exhibiting unique pathogenesis and immune evasions, thereby complicating its eradication. The treatment of MSKI typically necessitates aggressive debridement and the complete removal of infected implant, bone and soft tissues, followed by antibiotics therapy. Novel diagnostics such as ELISA, NGS or immunoassays may complement existing diagnostic accuracy and provide guidance for antibiotics therapy. Ongoing research is exploring emerging diagnostic transcriptomes and proteomes, further enhancing our understanding and approach to tackling MSKI.
Notes
Conflicts of interest
Irvin Oh is a editorial board member of the journal but was not involved in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflicts of interest relevant to this article were reported.
Funding
This study was supported by a grant from the Orthopaedic Research and Education Foundation (OREF) Career Development Grant number 21-064, and the National Institute of Health (NIH)/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant number R21AR073321 and R21AR074571, awarded to Irvin Oh, MD.
References
1. Nishitani K, Beck CA, Rosenberg AF, Kates SL, Schwarz EM, Daiss JL. A diagnostic serum antibody test for patients with Staphylococcus aureus osteomyelitis. Clin Orthop Relat Res. 2015; 473:2735–49.
2. Nishitani K, Sutipornpalangkul W, de Mesy Bentley KL, Varrone JJ, Bello-Irizarry SN, Ito H, et al. Quantifying the natural history of biofilm formation in vivo during the establishment of chronic implant-associated Staphylococcus aureus osteomyelitis in mice to identify critical pathogen and host factors. J Orthop Res. 2015; 33:1311–9.
3. Masters EA, Trombetta RP, de Mesy Bentley KL, Boyce BF, Gill AL, Gill SR, et al. Evolving concepts in bone infection: redefining “biofilm”, “acute vs. chronic osteomyelitis”, “the immune proteome” and “local antibiotic therapy”. Bone Res. 2019; 7:20.
4. Masters EA, Ricciardi BF, Bentley KL, Moriarty TF, Schwarz EM, Muthukrishnan G. Skeletal infections: microbial pathogenesis, immunity and clinical management. Nat Rev Microbiol. 2022; 20:385–400.
5. Longo M, Pennington Z, Gelfand Y, De la Garza Ramos R, Echt M, Ahmed AK, et al. Readmission after spinal epidural abscess management in urban populations: a bi-institutional study. J Neurosurg Spine. 2019; 32:465–72.
6. Beam E, Osmon D. Prosthetic joint infection update. Infect Dis Clin North Am. 2018; 32:843–59.
7. Oh I, Muthukrishnan G, Ninomiya MJ, Brodell JD Jr, Smith BL, Lee CC, et al. Tracking anti-Staphylococcus aureus antibodies produced in vivo and ex vivo during foot salvage therapy for diabetic foot infections reveals prognostic insights and evidence of diversified humoral immunity. Infect Immun. 2018; 86:e00629–18.
8. Lipof JS, Jones CM, Daiss J, Oh I. Comparative study of culture, next-generation sequencing, and immunoassay for identification of pathogen in diabetic foot ulcer. J Orthop Res. 2021; 39:2638–45.
9. Sulovari A, Ninomiya MJ, Beck CA, Ricciardi BF, Ketonis C, Mesfin A, et al. Clinical utilization of species-specific immunoassays for identification of Staphylococcus aureus and Streptococcus agalactiae in orthopedic infections. J Orthop Res. 2021; 39:2141–50.
10. Chaussade H, Uckay I, Vuagnat A, Druon J, Gras G, Rosset P, et al. Antibiotic therapy duration for prosthetic joint infections treated by debridement and implant retention (DAIR): similar long-term remission for 6 weeks as compared to 12 weeks. Int J Infect Dis. 2017; 63:37–42.
11. Li HK, Rombach I, Zambellas R, Walker AS, McNally MA, Atkins BL, et al. Oral versus intravenous antibiotics for bone and joint infection. N Engl J Med. 2019; 380:425–36.
12. Scarborough M, Li HK, Rombach I, Zambellas R, Walker AS, McNally M, et al. Oral versus intravenous antibiotics for bone and joint infections: the OVIVA non-inferiority RCT. Health Technol Assess. 2019; 23:1–92.
13. Blumberg G, Long B, Koyfman A. Clinical mimics: an emergency medicine-focused review of cellulitis mimics. J Emerg Med. 2017; 53:475–84.
14. Uy JP, Nuwayhid N, Saadeh C. Unusual presentations of gout: tips for accurate diagnosis. Postgrad Med. 1996; 100:253–68.
15. Lipsky BA, Senneville E, Abbas ZG, Aragon-Sanchez J, Diggle M, Embil JM, et al. Guidelines on the diagnosis and treatment of foot infection in persons with diabetes (IWGDF 2019 update). Diabetes Metab Res Rev. 2020; 36 Suppl 1:e3280.
16. Lu K, Zhang J, Cheng J, Liu H, Yang C, Yin L, et al. Incidence and risk factors for surgical site infection after open reduction and internal fixation of intra-articular fractures of distal femur: a multicentre study. Int Wound J. 2019; 16:473–8.
17. McDonald LS, Bavaro MF, Hofmeister EP, Kroonen LT. Hand infections. J Hand Surg Am. 2011; 36:1403–12.
18. Park KH, Kim DY, Lee YM, Lee MS, Kang KC, Lee JH, et al. Selection of an appropriate empiric antibiotic regimen in hematogenous vertebral osteomyelitis. PLoS One. 2019; 14:e0211888.
19. Ricciardi BF, Muthukrishnan G, Masters EA, Kaplan N, Daiss JL, Schwarz EM. New developments and future challenges in prevention, diagnosis, and treatment of prosthetic joint infection. J Orthop Res. 2020; 38:1423–35.
20. Holtfreter S, Kolata J, Broker BM. Towards the immune proteome of Staphylococcus aureus: the anti-S. aureus antibody response. Int J Med Microbiol. 2010; 300:176–92.
21. Lebon A, Labout JA, Verbrugh HA, Jaddoe VW, Hofman A, van Wamel W, et al. Dynamics and determinants of Staphylococcus aureus carriage in infancy: the Generation R Study. J Clin Microbiol. 2008; 46:3517–21.
22. Howden BP, Giulieri SG, Wong Fok Lung T, Baines SL, Sharkey LK, Lee JY, et al. Staphylococcus aureus host interactions and adaptation. Nat Rev Microbiol. 2023; 21:380–95.
23. Jenkins A, Diep BA, Mai TT, Vo NH, Warrener P, Suzich J, et al. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio. 2015; 6:e02272–14.
24. de Mesy Bentley KL, MacDonald A, Schwarz EM, Oh I. Chronic osteomyelitis with Staphylococcus aureus deformation in submicron canaliculi of osteocytes: a case report. JBJS Case Connect. 2018; 8:e8.
25. Guo H, Tong Y, Cheng J, Abbas Z, Li Z, Wang J, et al. Biofilm and small colony variants: an update on Staphylococcus aureus strategies toward drug resistance. Int J Mol Sci. 2022; 23:1241.
26. Muthukrishnan G, Masters EA, Daiss JL, Schwarz EM. Mechanisms of Immune evasion and bone tissue colonization that make Staphylococcus aureus the primary pathogen in osteomyelitis. Curr Osteoporos Rep. 2019; 17:395–404.
27. Hao SP, Masters EA, Ninomiya MJ, Beck CA, Schwarz EM, Daiss JL, et al. Species-specific immunoassay aids identification of pathogen and tracks infectivity in foot infection. Foot Ankle Int. 2021; 42:363–72.
28. Bennett MR, Dong J, Bombardi RG, Soto C, Parrington HM, Nargi RS, et al. Human VH1-69 gene-encoded human monoclonal antibodies against Staphylococcus aureus IsdB use at least three distinct modes of binding to inhibit bacterial growth and pathogenesis. mBio. 2019; 10:e02473–19.
29. Pishchany G, Sheldon JR, Dickson CF, Alam MT, Read TD, Gell DA, et al. IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus. J Infect Dis. 2014; 209:1764–72.
30. Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006; 188:8421–9.
31. Tsai CM, Caldera JR, Hajam IA, Chiang AW, Tsai CH, Li H, et al. Non-protective immune imprint underlies failure of Staphylococcus aureus IsdB vaccine. Cell Host Microbe. 2022; 30:1163–72.
32. McNeely TB, Shah NA, Fridman A, Joshi A, Hartzel JS, Keshari RS, et al. Mortality among recipients of the Merck V710 Staphylococcus aureus vaccine after postoperative S. aureus infections: an analysis of possible contributing host factors. Hum Vaccin Immunother. 2014; 10:3513–6.
33. Nishitani K, Ishikawa M, Morita Y, Yokogawa N, Xie C, de Mesy Bentley KL, et al. IsdB antibody-mediated sepsis following S. aureus surgical site infection. JCI Insight. 2020; 5:e141164.
34. Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012; 34:237–59.
35. Ahmad-Mansour N, Loubet P, Pouget C, Dunyach-Remy C, Sotto A, Lavigne JP, et al. Staphylococcus aureus toxins: an update on their pathogenic properties and potential treatments. Toxins (Basel). 2021; 13:677.
36. Spaan AN, van Strijp JA, Torres VJ. Leukocidins: Staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol. 2017; 15:435–47.
37. Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014; 12:49–62.
38. Brandt SL, Putnam NE, Cassat JE, Serezani CH. Innate immunity to Staphylococcus aureus: evolving paradigms in soft tissue and invasive infections. J Immunol. 2018; 200:3871–80.
39. Watson AR, Janik DK, Lee WT. Superantigen-induced CD4 memory T cell anergy. I. Staphylococcal enterotoxin B induces Fyn-mediated negative signaling. Cell Immunol. 2012; 276:16–25.
40. Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A. 2000; 97:5399–404.
41. Rooijakkers SH, van Wamel WJ, Ruyken M, van Kessel KP, van Strijp JA. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005; 7:476–84.
42. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010; 8:623–33.
43. Redlich K, Smolen JS. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012; 11:234–50.
44. Urish KL, DeMuth PW, Craft DW, Haider H, Davis CM 3rd. Pulse lavage is inadequate at removal of biofilm from the surface of total knee arthroplasty materials. J Arthroplasty. 2014; 29:1128–32.
45. Cheng AG, DeDent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011; 19:225–32.
46. Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 2010; 6:e1001036.
47. Kavanagh N, Ryan EJ, Widaa A, Sexton G, Fennell J, O’Rourke S, et al. Staphylococcal osteomyelitis: disease progression, treatment challenges, and future directions. Clin Microbiol Rev. 2018; 31:e00084–17.
48. Hofstee MI, Riool M, Terjajevs I, Thompson K, Stoddart MJ, Richards RG, et al. Three-dimensional in vitro Staphylococcus aureus abscess communities display antibiotic tolerance and protection from neutrophil clearance. Infect Immun. 2020; 88:e00293–20.
49. Farnsworth CW, Schott EM, Jensen SE, Zukoski J, Benvie AM, Refaai MA, et al. Adaptive upregulation of clumping factor A (ClfA) by Staphylococcus aureus in the obese, type 2 diabetic host mediates increased virulence. Infect Immun. 2017; 85:e01005–16.
50. de Mesy Bentley KL, Trombetta R, Nishitani K, Bello-Irizarry SN, Ninomiya M, Zhang L, et al. Evidence of Staphylococcus aureus deformation, proliferation, and migration in canaliculi of live cortical bone in murine models of osteomyelitis. J Bone Miner Res. 2017; 32:985–90.
51. Muthukrishnan G, Beck CA, Owen JR, Xie C, Kates SL, Daiss JL. Serum antibodies against Staphylococcus aureus can prognose treatment success in patients with bone infections. J Orthop Res. 2021; 39:2169–76.
52. Choi Y, Oda E, Waldman O, Sajda T, Beck C, Oh I. Next-generation sequencing for pathogen identification in infected foot ulcers. Foot Ankle Orthop. 2021; 6:24730114211026933.
53. Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, Hanssen AD, et al. Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin Infect Dis. 2018; 67:1333–8.
54. Namdari S, Nicholson T, Abboud J, Lazarus M, Ramsey ML, Williams G, et al. Comparative study of cultures and next-generation sequencing in the diagnosis of shoulder prosthetic joint infections. J Shoulder Elbow Surg. 2019; 28:1–8.
55. Tarabichi M, Shohat N, Goswami K, Alvand A, Silibovsky R, Belden K, et al. Diagnosis of periprosthetic joint infection: the potential of next-generation sequencing. J Bone Joint Surg Am. 2018; 100:147–54.
56. Tarabichi M, Shohat N, Goswami K, Parvizi J. Can next generation sequencing play a role in detecting pathogens in synovial fluid? Bone Joint J. 2018; 100-B:127–33.
57. Yang Y, Lin J, Guo S, Xue X, Wang Y, Qiu S, et al. RRM2 protects against ferroptosis and is a tumor biomarker for liver cancer. Cancer Cell Int. 2020; 20:587.
58. Tang B, Xu W, Wang Y, Zhu J, Wang H, Tu J, et al. Identification of critical ferroptosis regulators in lung adenocarcinoma that RRM2 facilitates tumor immune infiltration by inhibiting ferroptotic death. Clin Immunol. 2021; 232:108872.
Fig. 1.
Examples of musculoskeletal infections. (A) Diabetic foot infection, (B) implant-associated periprosthetic infection, (C) infected tenosynovitis of the flexor hallucis longus tendon, and (D) septic arthritis of the native ankle joint.
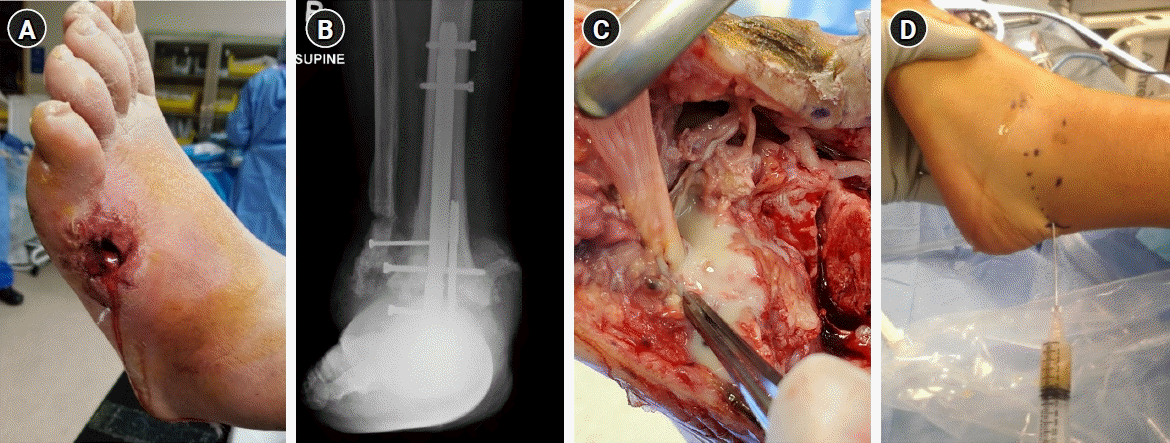
Fig. 2.
Staphylococcus aureus pathogenesis in musculoskeletal infections. Various pathogenic mechanisms employed by S. aureus. Osteocyte-lacuno canalicular network (OLCN) invasion permits S. aureus to evade immune cells while propagating along the submicron channel. Biofilm formation on implant surfaces or necrotic bone provides biologic shield, aggregation, and growth of the microorganisms with increase antibiotic-resistant gene transfers. The process of abscess formation involves the aggregation of S. aureus to establish a colony and the development of a fibrous pseudocapsule, confining immune cells to the periphery and restricting their access to the center where bacteria replication persists.
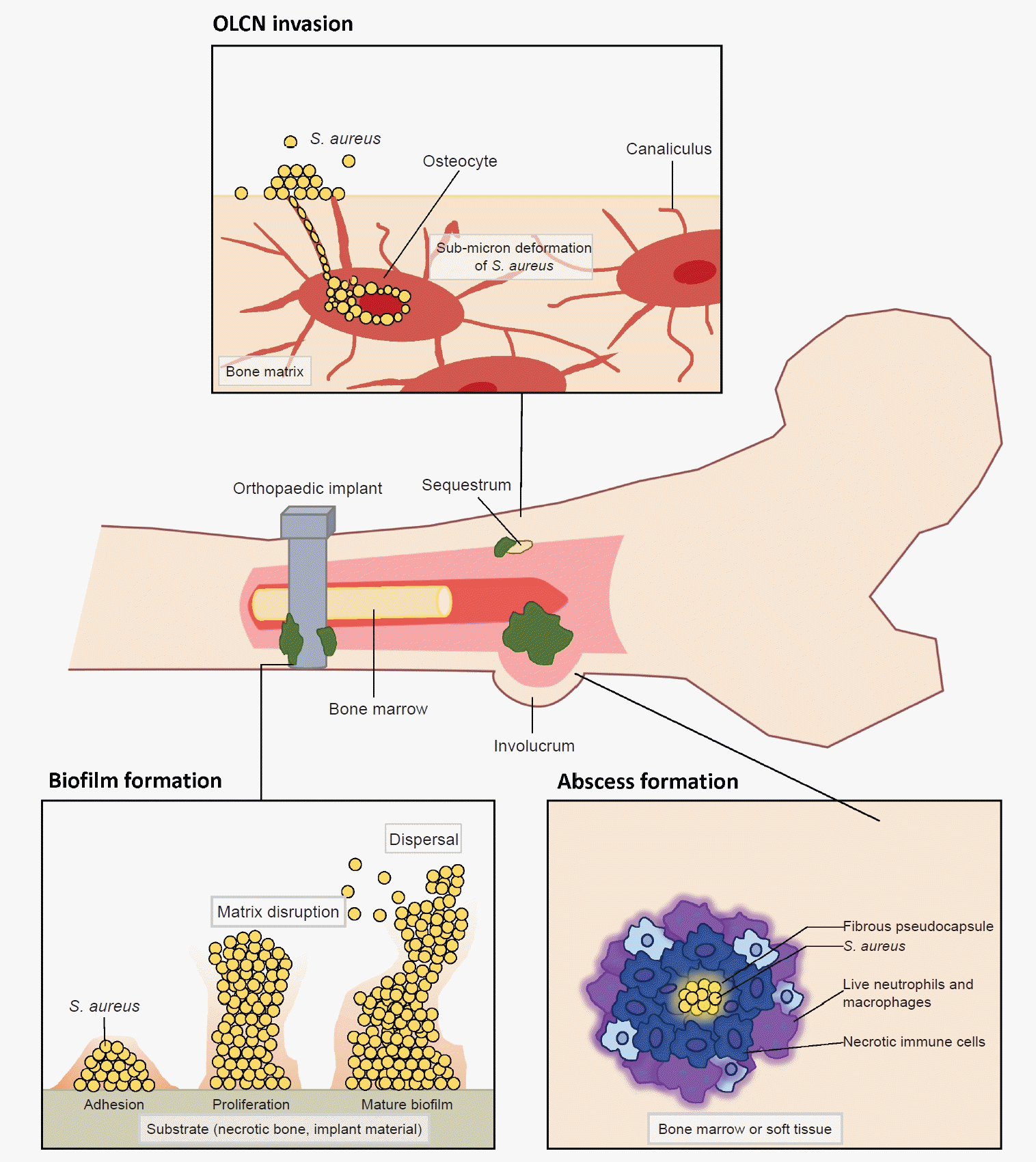
Fig. 3.
Staphylococcus aureus iron-regulated surface determinant (Isd) system. S. aureus utilizes the Isd system to strip iron from the host’s hemoglobin. IsdH, IsdB, and IsdA are anchored to the cell wall. Hemoglobin is captured by IsdH and IsdB through near iron transport (NEAT) domains. IsdB has two NEAT domains: NEAT1 binds to hemoglobin, and NEAT2 binds and transports the heme. The IsdB removes heme from hemoglobin and transports it to IsdA, which subsequently transfers the heme to the intracellular IsdC protein. The heme is degraded within the cell so that iron is released for utilization by S. aureus.
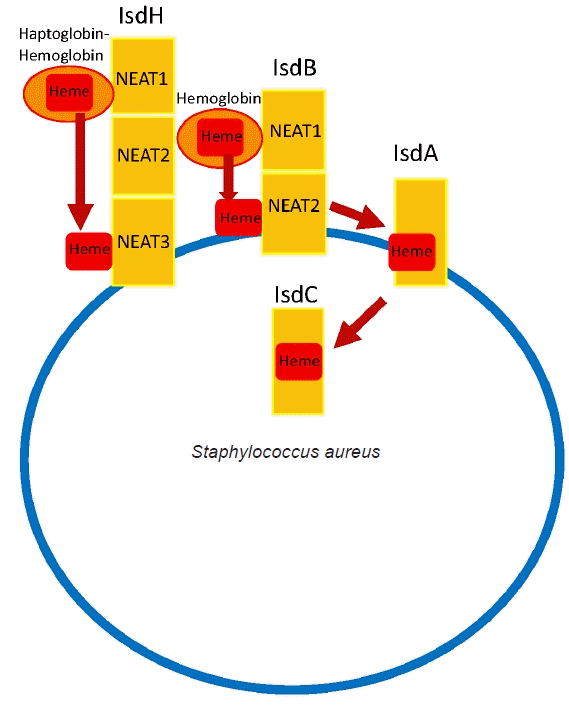
Fig. 4.
Staphylococcus aureus immune evasion mechanisms. S. aureus secretes toxins—α-hemolysin (Hla), β-hemolysin, γ-hemolysin (HlgAB and HlgCB), leucocidin A/B (LukAB) and Panton-Valentine leukocidin (PVL)—that directly destroy neutrophils, macrophages and other antigen-presenting cells. S. aureus enterotoxin B and C (SEB, SEC) and toxic shock syndrome toxin 1 (TSST1) induce a shift in M2 macrophage polarization and promote formation of myeloid-derived suppressor cells. S. aureus utilizes microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) to facilitate its internal invasion of macrophages and neutrophils (ClfA, ClfB, fibronectin-binding protein A [FnBPA], fibronectin-binding protein B [FnBPB]). S. aureus also interferes with complement-mediated opsonization and phagocytosis through chemotaxis inhibitory protein of S. aureus (CHIPS), Staphylococcal complement inhibitor (SCIN), coagulase (CoA), and extracellular fibrinogen binding protein (Efb). SpA binds certain antibodies, blocking antibody-mediated phagocytosis and concurrently initiating proliferative B cell apoptosis. Staphylokinase (Sak) can directly degrade IgG.
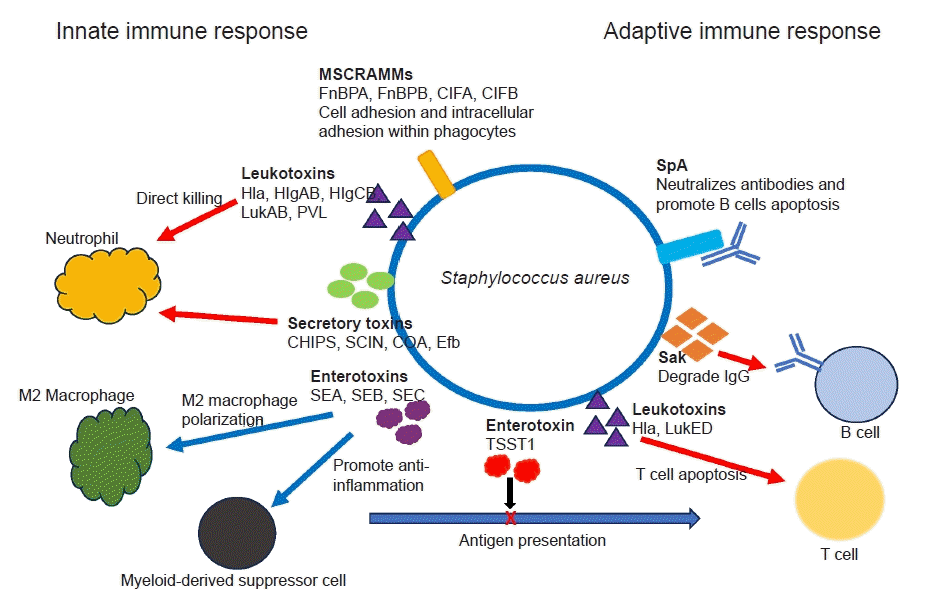
Fig. 5.
Transmission electron microscopy (TEM) evidence of Staphylococcus aureus invasion of the osteocyte-lacuno canalicular network. TEM investigation of an infected bone sample from a diabetic foot osteomyelitis patient demonstrated Staphylococcus deformation and invasion of the submicron osteocytic-canalicular system. Standard culture of the infected bone sample identified S. aureus. (A) TEM image of a dead osteocyte and its canaliculus colonized by deformed rod-shaped bacteria. (B) Higher-magnification TEM image show submicron-sized deformed and elongated bacteria [24].
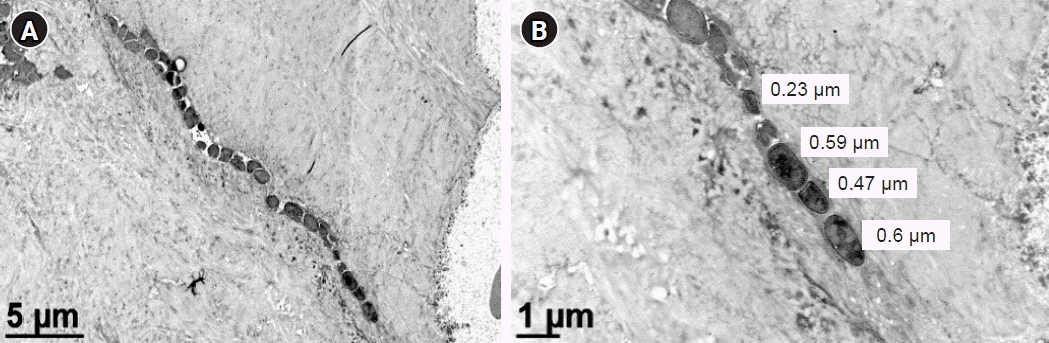
Fig. 6.
Development of novel diagnostics for musculoskeletal infections. Blood samples are obtained from patients with musculoskeletal infections (MSKI) to isolate peripheral blood mononuclear cells for transcriptome analysis. Upregulated ribonucleotide reductase regulatory subunit M2 (RRM2), a transcription factor in the Myb-related protein B (MYBL2), and a neutrophil-specific cell surface glycoprotein (CD177) have been detected, suggesting their potential role as biomarkers for MSKI diagnosis. Additionally, species-specific serum immunoassays are being explored to detect Staphylococcus aureus infection. Concurrently, research is underway on serum proteomics for the identification of protein biomarkers associated with MSKI. Isd, iron-regulated surface determinant; SCIN, Staphylococcal complement inhibitor; PBMC, peripheral blood mononuclear cell.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download